The membrane that encases a organic cell just isn’t merely a barrier; it’s chock stuffed with proteins concerned in all types of crucial organic features. To actually perceive what membrane proteins are doing and the way, researchers must understand how they’re organized and the way they work together with each other. However uncovering that data is difficult.
Yale researchers have now developed a brand new microscopy methodology known as Native-nanoBleach that overcomes the principle challenges to understanding membrane protein group, together with the issue in finding out these membranes with out disrupting the native atmosphere and limits to the decision of sunshine microscopes usually used to review them.
And to reveal the effectiveness of the brand new methodology, they efficiently utilized it to a organic conundrum—pertaining to proteins concerned within the growth of pancreatic cancers and the way they could be focused for therapy—that has remained unsolved for many years.
They describe the brand new methodology and its benefits in a brand new research revealed in Nature Nanotechnology.
Strategies usually used within the research of membrane protein group require eradicating the native membrane atmosphere surrounding proteins after which placing remoted proteins of curiosity into environments that mimic however don’t totally replicate the complexity of the true cell membrane, mentioned Moitrayee Bhattacharyya, assistant professor of pharmacology at Yale College of Medication and senior creator of the research. This strategy, Bhattacharyya mentioned, removes essential context for the reason that proteins work together with the molecules that encompass them.
Second, gentle microscopes, that are generally used to look at protein group, do not have the decision wanted to find out whether or not proteins close to one another are actually interacting or are merely neighbors on membranes.
Final, the quantity of a specific protein present in a cell membrane could also be too low or too excessive for present strategies of research. In these situations, researchers need to make changes; they both have to copy proteins that of their pure state are too few in quantity or separate out proteins from samples the place there are too many. However this once more can take away essential context in regards to the pure state of proteins as they sit and performance in cell membranes.
“Ideally, we might have a way that may work with any endogenous degree of cell membrane protein expression,” mentioned Bhattacharyya.
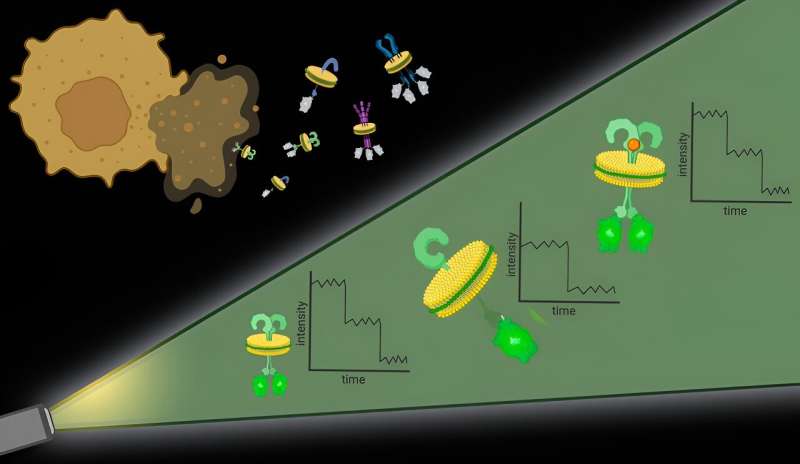
To sort out the primary problem, the researchers used specific molecules, forms of polymers, to primarily punch out protein complexes with their surrounding cell membrane intact. “It is just like the cell membrane is a sheet of cookie dough and the polymers are cookie cutters,” mentioned Bhattacharyya.
These bits of protein with surrounding cell membrane, dubbed native nanodiscs, are roughly 10 nanometers in diameter, sufficiently small that any proteins contained within the nanodisc are doubtless interacting, which addresses the second problem. Additional, this strategy works with any cell membrane protein in any quantity, permitting researchers to look at proteins at their pure ranges in native membranes.
As soon as the nanodiscs are generated, researchers can use any variety of generally used methods to hone in on a specific protein of curiosity. They then quantify the proteins in every nanodisc with the assistance of fluorescent molecules connected to them.
It is an strategy that provides excessive spatial decision with out the necessity for specialised {hardware}, mentioned Bhattacharyya.
“This work presents a brand new approach to grasp how membrane proteins—which characterize about 60% of drug targets—assemble into useful items on or throughout the native lipid bilayer,” mentioned Gerard Walker, co-first creator of the paper and a graduate scholar in Bhattacharyya’s lab.
To reveal how this methodology could be utilized, the researchers took on a decades-long debate in biology. A protein known as KRas is mutated in additional than 90% of human pancreatic cancers, sparking immense medical and therapeutic curiosity. Whether or not KRas subunits come collectively to kind dimers (two items) or oligomers (greater than two items) on cell membranes has remained the main target of longstanding investigation.
Research, nonetheless, have produced conflicting findings. Animal and mobile research, which lack detailed molecular decision, present proof that KRas items come collectively on cell membranes. In the meantime, biophysical analyses, which don’t retain the native membrane round proteins, have discovered KRas stays in single items, or monomers.
“With our methodology, we get the very best of each worlds,” mentioned Bhattacharyya. “We retain the native membrane atmosphere and we’ve very excessive spatial and single-molecule decision. After we utilized our methodology, we discovered that KRas exists as dimers and monomers in related quantities. However when KRas is mutated, as in pancreatic cancers, dimers enhance and monomers lower.”
The discovering highlights the significance of the native cell membrane for understanding membrane proteins and identifies a goal—decreasing KRas dimerization—for most cancers therapy. This is only one of some ways this methodology could possibly be used to grasp the function of membrane protein group in illness, Bhattacharyya mentioned.
“It is actually rewarding to see Native-nanoBleach already being efficiently utilized to all kinds of urgent organic questions within the Bhattacharyya lab and past,” mentioned Caroline Brown, co-first creator of the research and a Ph.D. candidate within the lab of co-author Kallol Gupta, assistant professor of cell biology.
Membrane proteins make up a 3rd of all the proteins within the human physique and this strategy can be utilized to review any of them, mentioned Bhattacharyya.
“It is a normal approach,” she mentioned. “There’s actually no limitation.”
Going ahead, Bhattacharyya and her colleagues hope to increase this strategy to finding out protein group within the membranes of assorted organelles, buildings, like mitochondria, which are contained inside cells.
Extra data:
Gerard Walker et al, Oligomeric group of membrane proteins from native membranes at nanoscale spatial and single-molecule decision, Nature Nanotechnology (2023). DOI: 10.1038/s41565-023-01547-4
Offered by
Yale College
Quotation:
Utilizing molecular ‘cookie cutters’ to view membrane protein group (2023, December 21)
retrieved 23 December 2023
from https://phys.org/information/2023-12-molecular-cookie-cutters-view-membrane.html
This doc is topic to copyright. Other than any honest dealing for the aim of personal research or analysis, no
half could also be reproduced with out the written permission. The content material is supplied for data functions solely.


