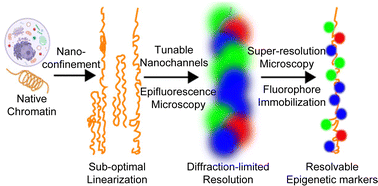
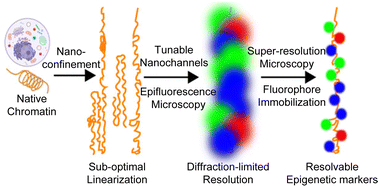
Nanofluidic linearization and optical mapping of bare DNA have been reported within the analysis literature, and carried out in industrial devices. Nevertheless, the decision with which DNA options may be resolved remains to be inherently restricted by each Brownian movement and diffraction-limited optics. Direct evaluation of native chromatin is additional hampered by issue in electrophoretic manipulation, which is routinely used for DNA evaluation. This paper describes the event of a three-layer, tunable, nanochannel system that permits non-electrophoretic linearization and immobilization of native chromatin. Moreover, by means of cautious collection of self-blinking fluorescent dyes and the design of the nanochannel system, we obtain direct stochastic optical reconstruction microscopy (dSTORM) super-resolution imaging of the linearized chromatin. As an preliminary demonstration, rDNA chromatin extracted from Tetrahymena is analyzed by multi-color imaging of complete DNA, newly synthesized DNA, and newly synthesized histone H3. Our evaluation reveals a comparatively even distribution of newly synthesized H3 throughout two halves of the rDNA chromatin with palindromic symmetry, supporting dispersive nucleosome segregation. As a proof-of-concept research, our work achieves super-resolution imaging of native chromatin fibers linearized and immobilized in tunable nanochannels. It opens up a brand new avenue for amassing long-range and high-resolution epigenetic data in addition to genetic data.