Jun 28, 2023
(Nanowerk Information) A crew of researchers led by Feng Zhang on the Broad Institute of MIT and Harvard and the McGovern Institute for Mind Analysis at MIT has uncovered the primary programmable RNA-guided system in eukaryotes — organisms that embrace fungi, crops, and animals.
In a research in Nature (“Fanzor is a eukaryotic programmable RNA-guided endonuclease”), the crew describes how the system is predicated on a protein referred to as Fanzor. They confirmed that Fanzor proteins use RNA as a information to focus on DNA exactly, and that Fanzors will be reprogrammed to edit the genome of human cells. The compact Fanzor programs have the potential to be extra simply delivered to cells and tissues as therapeutics than CRISPR/Cas programs, and additional refinements to enhance their focusing on effectivity might make them a worthwhile new expertise for human genome enhancing.
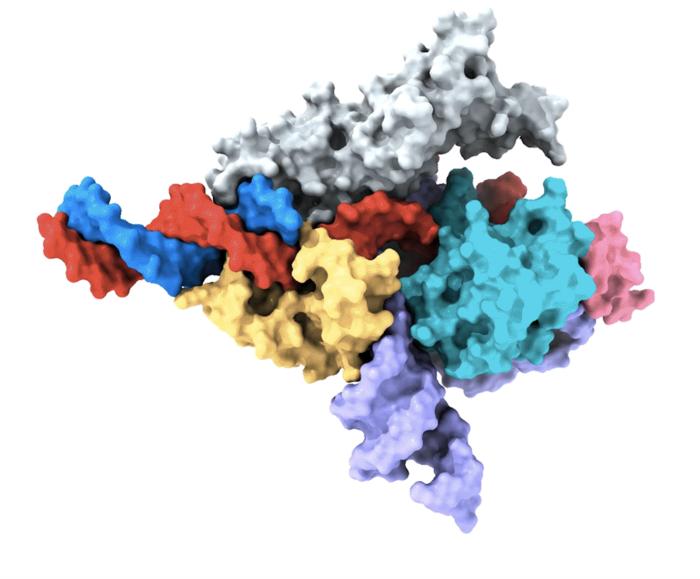 A Cryo-EM map of a Fanzor protein (grey, yellow, mild blue, and pink) in advanced with ωRNA (purple) and its goal DNA (crimson). Non-target DNA strand in blue. (Picture: Zhang lab, Broad Institute of MIT and Harvard/McGovern Institute for Mind Analysis at MIT)
CRISPR/Cas was first found in prokaryotes (micro organism and different single-cell organisms that lack nuclei) and scientists together with Zhang’s lab have lengthy questioned whether or not comparable programs exist in eukaryotes. The brand new research demonstrates that RNA-guided DNA-cutting mechanisms are current throughout all kingdoms of life.
“CRISPR-based programs are broadly used and highly effective as a result of they are often simply reprogrammed to focus on completely different websites within the genome,” stated Zhang, senior creator on the research and a core institute member on the Broad, an investigator at MIT’s McGovern Institute, the James and Patricia Poitras Professor of Neuroscience at MIT, and a Howard Hughes Medical Institute investigator. “This new system is one other strategy to make exact adjustments in human cells, complementing the genome enhancing instruments we have already got.”
A Cryo-EM map of a Fanzor protein (grey, yellow, mild blue, and pink) in advanced with ωRNA (purple) and its goal DNA (crimson). Non-target DNA strand in blue. (Picture: Zhang lab, Broad Institute of MIT and Harvard/McGovern Institute for Mind Analysis at MIT)
CRISPR/Cas was first found in prokaryotes (micro organism and different single-cell organisms that lack nuclei) and scientists together with Zhang’s lab have lengthy questioned whether or not comparable programs exist in eukaryotes. The brand new research demonstrates that RNA-guided DNA-cutting mechanisms are current throughout all kingdoms of life.
“CRISPR-based programs are broadly used and highly effective as a result of they are often simply reprogrammed to focus on completely different websites within the genome,” stated Zhang, senior creator on the research and a core institute member on the Broad, an investigator at MIT’s McGovern Institute, the James and Patricia Poitras Professor of Neuroscience at MIT, and a Howard Hughes Medical Institute investigator. “This new system is one other strategy to make exact adjustments in human cells, complementing the genome enhancing instruments we have already got.”
 A Cryo-EM map of a Fanzor protein (grey, yellow, mild blue, and pink) in advanced with ωRNA (purple) and its goal DNA (crimson). Non-target DNA strand in blue. (Picture: Zhang lab, Broad Institute of MIT and Harvard/McGovern Institute for Mind Analysis at MIT)
CRISPR/Cas was first found in prokaryotes (micro organism and different single-cell organisms that lack nuclei) and scientists together with Zhang’s lab have lengthy questioned whether or not comparable programs exist in eukaryotes. The brand new research demonstrates that RNA-guided DNA-cutting mechanisms are current throughout all kingdoms of life.
“CRISPR-based programs are broadly used and highly effective as a result of they are often simply reprogrammed to focus on completely different websites within the genome,” stated Zhang, senior creator on the research and a core institute member on the Broad, an investigator at MIT’s McGovern Institute, the James and Patricia Poitras Professor of Neuroscience at MIT, and a Howard Hughes Medical Institute investigator. “This new system is one other strategy to make exact adjustments in human cells, complementing the genome enhancing instruments we have already got.”
A Cryo-EM map of a Fanzor protein (grey, yellow, mild blue, and pink) in advanced with ωRNA (purple) and its goal DNA (crimson). Non-target DNA strand in blue. (Picture: Zhang lab, Broad Institute of MIT and Harvard/McGovern Institute for Mind Analysis at MIT)
CRISPR/Cas was first found in prokaryotes (micro organism and different single-cell organisms that lack nuclei) and scientists together with Zhang’s lab have lengthy questioned whether or not comparable programs exist in eukaryotes. The brand new research demonstrates that RNA-guided DNA-cutting mechanisms are current throughout all kingdoms of life.
“CRISPR-based programs are broadly used and highly effective as a result of they are often simply reprogrammed to focus on completely different websites within the genome,” stated Zhang, senior creator on the research and a core institute member on the Broad, an investigator at MIT’s McGovern Institute, the James and Patricia Poitras Professor of Neuroscience at MIT, and a Howard Hughes Medical Institute investigator. “This new system is one other strategy to make exact adjustments in human cells, complementing the genome enhancing instruments we have already got.”


