Identification and expression sample of the novel protein NbCRVP
The open studying body sequence of a cysteine-rich venom protein in N. benthamiana was amplified utilizing primers CRVPF/CRVPR (Extra file 2: Desk S1) for the primary time and named NbCRVP basing on the referenced genome of N. benthamiana (https://solgenomics.web/organism/Nicotiana_benthamiana/genome, accessed on Could 20, 2020). SMART evaluation (http://sensible.embl-heidelberg.de/, accessed on February 5, 2020) revealed that the obtained NbCRVP sequence contained an SCP area, which is the CAP area shared by all members of the CAP superfamily. Monitoring NbCRVP expression beneath viral an infection confirmed that NbCRVP expression was induced at 1 day post-viral inoculation (dpi) and reached its peak at 5 dpi (Extra file 1: Fig. S1a). Furthermore, NbCRVP expression was detected in roots, stems, leaves, and flowers of the wild-type N. benthamiana, with the very best expression stage discovered within the flower tissues (Extra file 1: Fig. S1b). Laser confocal statement of the subcellular localization of NbCRVP confirmed that the protein was distributed in all the cell define, however not within the nucleus. (Extra file 1: Figs. S1e, S2). Spraying 0.5 mM SA, 0.1 mM Me-JA, and 0.05 mM ethephon all upregulated NbCRVP expression by 4.15, 4.02, and 9.05 folds, respectively (Extra file 1: Fig. S1c). Against this, silencing the important thing genes NPR1, COI1, and EIN2 of SA, JA and ET signaling pathways confirmed solely TRV::NPR1 downregulated NbCRVP expression, with a 27% discount (Extra file 1: Fig. S1d). These outcomes recommend that NbCRVP could reply to viral an infection.
Silencing NbCRVP up-regulates the sensitivity of N. benthamiana to a number of RNA viruses
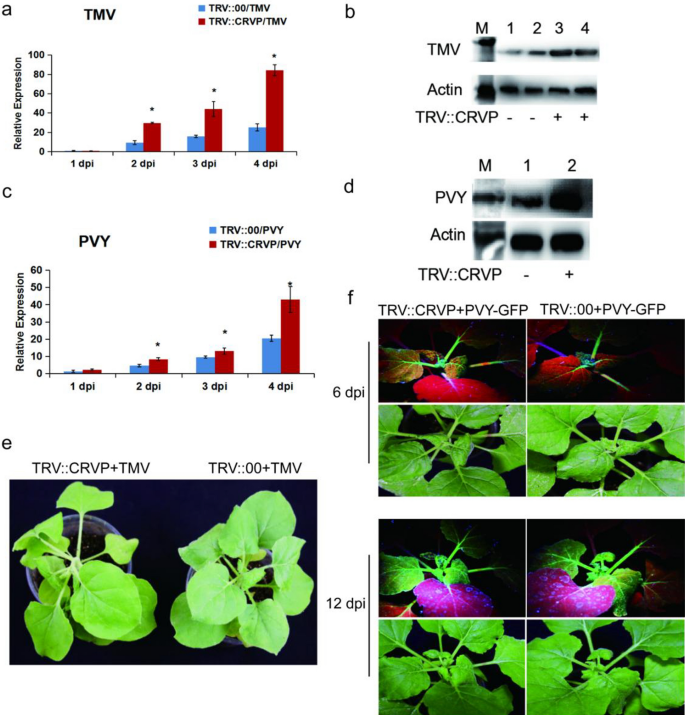
To make clear the perform of NbCRVP, VIGS know-how and viral inoculation experiment have been used to establish its organic perform from the expression distinction of viral CP on the gene and protein ranges. Firstly, the silencing effectivity of NbCRVP was detected at 14 days after infiltration of pTRV vectors (Extra file 1: Fig. S3), the virus was innoculated at 15 days after which the impact of silencing NbCRVP on viral an infection was detected. RT-qPCR information confirmed that the expression of TMV CP mRNA was considerably larger than that of the management group after silencing NbCRVP, which have been 2.44 instances and 1.46 instances at 2 and three dpi respectively (Fig. 2a). WB outcomes additionally confirmed that the TMV CP content material in TRV::CRVP group was larger than that within the management group (TRV::00) at 3 dpi (Fig. 2b). The organic signs of viral an infection within the remedy group have been additionally considerably extra extreme than these within the management group, and the remedy group confirmed extra extreme signs of necrosis of veins and lodging at 4 dpi (Fig. 2e). All these outcomes confirmed that silencing NbCRVP promoted TMV an infection to vegetation.
Silencing NbCRVP promotes each TMV and PVY an infection in N. benthamiana. a Impact of silencing NbCRVP on TMV an infection detected by RT-qPCR at 1, 2, 3, and 4 dpi. b Impact of silencing NbCRVP on TMV an infection on the protein stage. c Impact of silencing NbCRVP on PVY an infection detected by RT-qPCR at 1, 2, 3, and 4 dpi. d Impact of silencing NbCRVP on PVY an infection at protein ranges. e Results of silencing NbCRVP on organic signs of TMV an infection. f Results of silencing NbCRVP on GFP alerts after PVY-GFP an infection. All information have been analyzed utilizing the impartial pattern t take a look at. * signifies vital variations between two completely different therapies (P < 0.05)
The PVY-GFP inoculation assay additionally detected the degrees of PVY CP RNA and protein ranges in TRV::CRVP and TRV::00 teams. On the mRNA stage, the CP content material of the remedy group was 1.78, 1.36, and a pair of.10 instances that of the management group at 2, 3 and 4 dpi, respectively (Fig. 2c). At 4 dpi, the PVY CP antibody imprinted a stronger immune sign in TRV::CRVP samples (Fig. 2d). Furthermore, the GFP sign of the remedy group was additionally stronger than that of the management group at 6 dpi and 12 dpi (Fig. 2f). These outcomes displayed that silencing NbCRVP promoted PVY an infection to vegetation. Taken collectively, silencing NbCRVP up-regulated the sensitivity of N. benthamiana to a number of RNA viruses.
Overexpression of NbCRVP enhances the resistance of N. benthamiana to RNA viruses
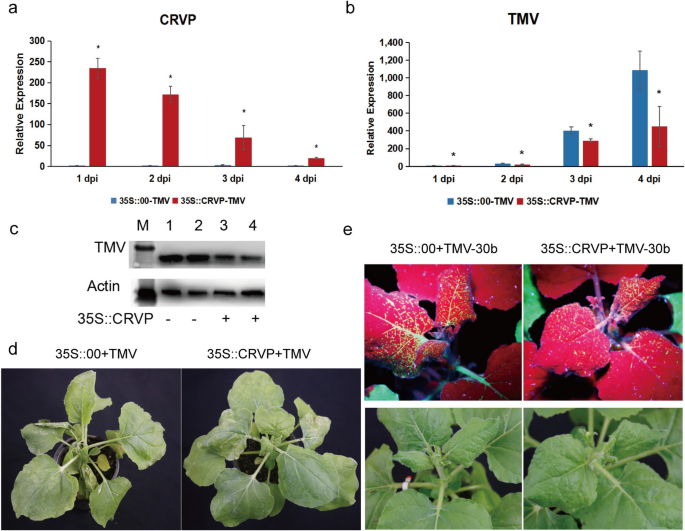
To additional verify the antiviral perform of NbCRVP, a transient overexpression experiment was carried out utilizing an overexpression vector with 35S promoter. The expression stage of NbCRVP was detected at 1, 2, 3, and 4 days after transient infiltration of p35S::CRVP-RFP A. tumefaciens. The outcomes confirmed that NbCRVP expression stage within the 35S::CRVP remedy group innoculated with TMV was 230.39, 69.48, 20.22, and 30.74 instances that of the 35S::00 management group additionally innoculated with TMV, respectively (Fig. 3a). The qRT-PCR outcomes confirmed that the TMV CP mRNA stage was decreased by 43%, 28% and 59% at 2, 3, and 4 dpi, respectively, within the 35S::CRVP-TMV group in contrast with the management group (35S::00-TMV) (Fig. 3b). The WB outcomes additional confirmed that TMV CP protein stage within the 35S::CRVP-TMV group was decrease than that within the management group at 3 dpi (Fig. 3c). Moreover, the management group exhibited typical signs of vein necrosis and wilting, whereas the remedy group confirmed no such apparent signs (Fig. 3d). Inoculation experiments with TMV-30b visually demonstrated a weaker GFP sign within the 35S::CRVP remedy group (Fig. 3e). These outcomes indicated that NbCRVP overexpression inhibited TMV an infection to N. benthamiana.
Overexpression of NbCRVP inhibits TMV an infection to N. benthamiana. a NbCRVP ranges after its overexpression. b TMV CP mRNA ranges after NbCRVP overexpression at 1, 2, 3, and 4 dpi. c Impact of NbCRVP overexpression on TMV an infection on the protein stage. d Impact of NbCRVP overexpression on organic signs of TMV an infection. e Impact of NbCRVP overexpression on GFP alerts after TMV-30b an infection. The info have been analyzed utilizing the impartial pattern t take a look at, and * signifies vital variations between the 2 completely different therapies (P < 0.05)
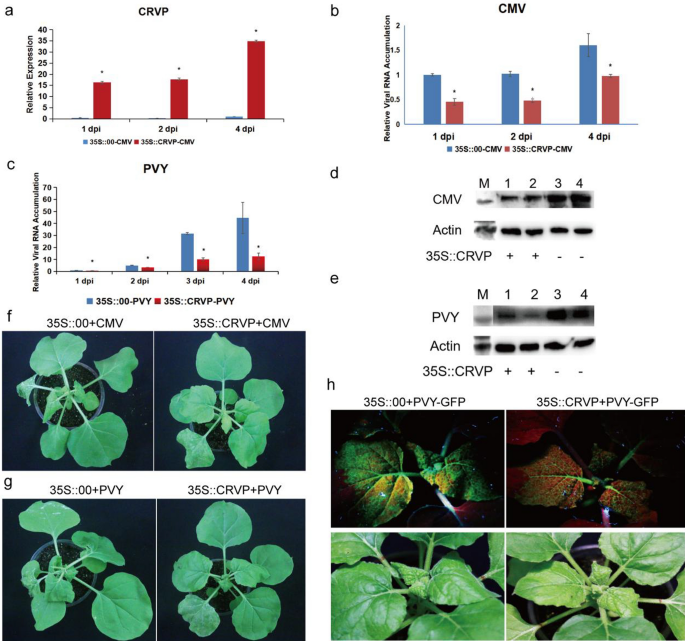
The expression ranges of NbCRVP within the transient NbCRVP overexpression group have been 86.11, 71, and 34.88 instances that of the management group at 1, 2, and 4 dpi, respectively (Fig. 4a). Nonetheless, the CMV CP ranges confirmed an reverse pattern, exhibiting 54%, 53%, and 39% lower at 1, 2, and 4 dpi, respectively, within the 35S::CRVP group in contrast with the 35S::00 group, which each inoculated with CMV (Fig. 4b). CMV CP ranges have been additionally decrease within the remedy group than within the management group at 4 dpi (Fig. 4d). In contrast with the 35S::CRVP remedy group inoculated with CMV, the 35S::00 management group inoculated with CMV confirmed extra apparent signs of virus an infection at 9 dpi (Fig. 4f). These outcomes additionally indicated that NbCRVP overexpression considerably inhibited CMV an infection to N. benthamiana.
NbCRVP overexpression inhibits CMV and PVY an infection to N. benthamiana. a NbCRVP ranges after its overexpression. b CMV CP mRNA ranges after NbCRVP overexpression at 1, 2, and 4 dpi measured by RT-qPCR. c PVY CP mRNA ranges after NbCRVP overexpression at 1, 2, 3, and 4 dpi, as measured by RT-qPCR. d Impact of NbCRVP overexpression on CMV an infection on the protein stage. e Impact of NbCRVP overexpression on PVY an infection on the protein stage. f Impact of NbCRVP overexpression on organic signs after CMV an infection. g Impact of NbCRVP overexpression on organic signs after PVY an infection. h Impact of NbCRVP overexpression on GFP alerts after PVY-GFP an infection. The info have been analyzed utilizing the impartial pattern t take a look at, and * signifies vital variations between the 2 completely different therapies (P < 0.05)
The relative expression ranges of NbCRVP and PVY CP in 35S::CRVP-PVY and 35S::00-PVY teams have been additionally detected after PVY-GFP an infection of N. benthamiana for 1, 2, 3, and 4 days (Fig. 4c). The PVY CP mRNA ranges have been decreased by 32%, 68%, and 72% within the 35S::CRVP-PVY group at 2, 3, 4 dpi, respectively, when put next with the 35S::00-PVY group (Fig. 4c). The PVY CP ranges have been in line with the outcomes at 4 dpi, proven in Fig. 4e. In contrast with the 35S::CRVP remedy group inoculated with PVY, the 35S::00 management group inoculated with PVY confirmed extra apparent signs of viral an infection at 9 dpi (Fig. 4g), weaker GFP alerts have been visualized within the 35S::CRVP remedy group than within the management group at 10 dpi, which each contaminated PVY-GFP (Fig. 4h). These outcomes additional illustrated that NbCRVP overexpression had a sure inhibitory impact on PVY an infection in N. benthamiana.
To extra comprehensively confirm the antiviral perform of NbCRVP, a multi-infection experiment of TMV, PVY, and CMV was carried out. The expression ranges of NbCRVP within the 35S::CRVP-virus group have been 3022.71, 52.21, 4.99, and 1.83 instances that of the 35S::00-virus group at 1, 2, 3, and 4 days after NbCRVP overexpression, respectively (Extra file 1: Fig. S4a). The RT-qPCR outcomes confirmed that TMV CP mRNA stage was decreased by 87%, 85%, 61%, and 58% within the remedy group in contrast with the management group at 1, 2, 3, and 4 dpi, respectively (Extra file 1: Fig. S4b). PVY CP mRNA stage was additionally decreased by 78%, 96%, and 89% within the remedy group in contrast with the management group at 1, 2, and 4 dpi, respectively (Extra file 1: Fig. S4b). Equally, CMV CP mRNA stage was decreased by 98%, 83%, 92%, and 38% within the remedy group in contrast with the management group at 1, 2, 3, and 4 dpi, respectively (Extra file 1: Fig. S4b). Furthermore, organic signs of viral an infection have been extra apparent within the 35S::0-virus group than within the management group (Extra file 1: Fig. S4c). The above outcomes clearly indicated that overexpression of NbCRVP made Nicotiana benthamiana much less vulnerable to RNA virus an infection, implying that NbCRVP is a novel protein enhancing the antiviral results in vegetation.
NbCRVP and NbCalB work together to exert enhanced antiviral results
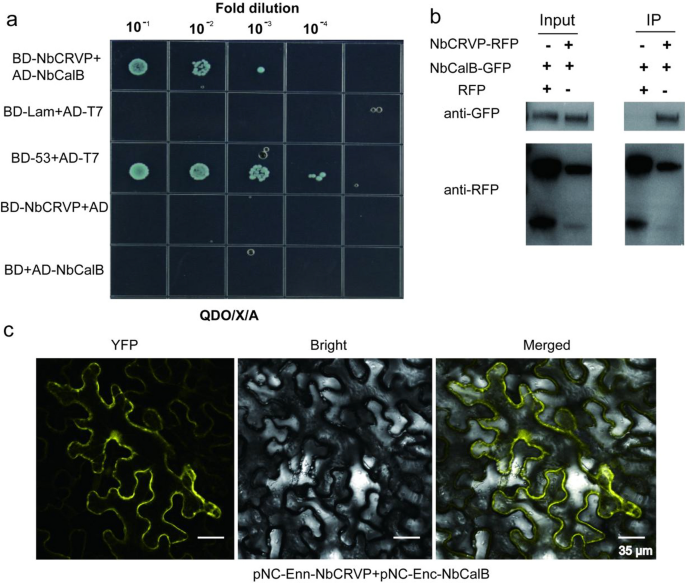
To additional discover the resistance mechanism of NbCRVP, a Y2H screening take a look at was carried out utilizing NbCRVP because the bait to establish potential interacting proteins. As proven in Extra file 1: Fig. S5, a powerful interplay was noticed between NbCalB and NbCRVP. The Y2H point-to-point verification outcomes confirmed that NbCRVP and NbCalB co-transformed strains grew on the SD/-Ade/-His/-Leu/-Trp/X/A (QDO/X/A) plates (Fig. 5a), whereas each NbCRVP and NbCalB confirmed no self-activation, indicating an interplay between them (Fig. 5a). The full plant proteins have been blotted utilizing an anti-GFP antibody and an anti-RFP antibody, respectively, which imprinted NbCalB-GFP and NbCRVP-RFP bands in each management and remedy teams (Fig. 5b Enter), indicating that each NbCalB-GFP and NbCRVP-RFP have been expressed. The full plant protein was purified utilizing the RFP magnetic beads, and co-IP outcomes confirmed that the anti-GFP antibody imprinted the NbCalB-GFP band within the remedy group however not within the management group (Fig. 5b IP). Against this, the anti-RFP antibody imprinted NbCRVP-RFP bands in each teams (Fig. 5b IP), confirming the interplay between NbCRVP and NbCalB. Furthermore, BIFC outcomes confirmed a powerful YFP sign, additional demonstrating an interplay between NbCRVP and NbCalB (Fig. 5c).
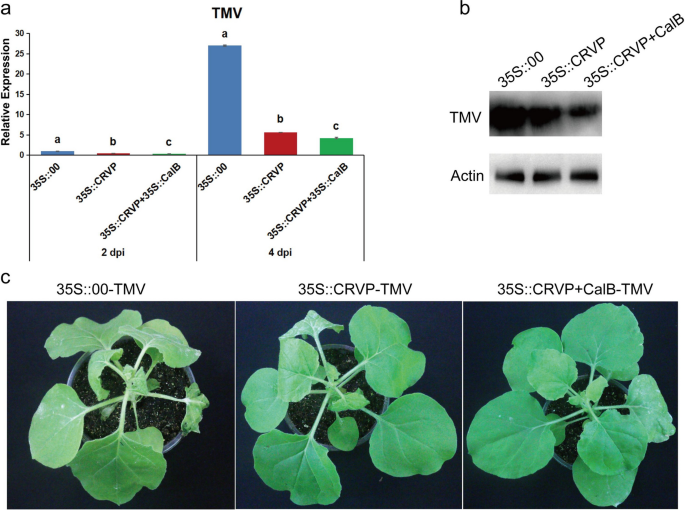
To determine the affect of the interplay between NbCRVP and NbCalB on viral an infection, A. tumefaciens have been reworked with expression vectors 35S::00, 35S::CRVP, or 35S::CRVP + 35S::CalB and contaminated with TMV. RT-qPCR outcomes confirmed that TMV CP mRNA ranges have been down-regulated in 35S::CRVP and 35S::CRVP + 35S::CalB reworked A. tumefaciens by 53% and 65% at 2 dpi, and 79% and 84% at 4 dpi, respectively, in contrast with that of 35S::00-transformed management A. tumefaciens (Fig. 6a). Western blotting outcomes for TMV CP protein ranges have been in line with the adjustments in mRNA ranges at 4 dpi (Fig. 6b). The management group 35S::00 inoculated with TMV confirmed apparent signs of vein necrosis and wilting, and the highest new leaves of the 35S::CRVP handled group confirmed curling signs. Nonetheless, the viral an infection signs of 35S::CRVP + CalB group have been the least noticeable (Fig. 6c). These outcomes indicated that NbCRVP and NbCalB work together to exert enhanced antiviral results.
A. tumefaciens-mediated co-expression of NbCRVP and NbCalB enhances the antiviral impact. a Results of 35S::CRVP and 35S::CalB co-expression on TMV an infection at 2 and 4 dpi, as measured by RT-qPCR, with 35S::00 and 35S::CRVP because the management therapies. The info have been analyzed by Duncan’s a number of vary exams within the ANOVA program of SPSS. Totally different letters point out vital variations among the many 4 remedy teams (P < 0.05). b Results of 35S::CRVP and 35S::CalB co-expression on TMV an infection at 4 dpi, as measured utilizing WB. c Results of 35S::CRVP and 35S::CalB co-expression on organic signs between the three completely different therapies
Preparation and characterization of PAMAM@pDNA nanocomposites
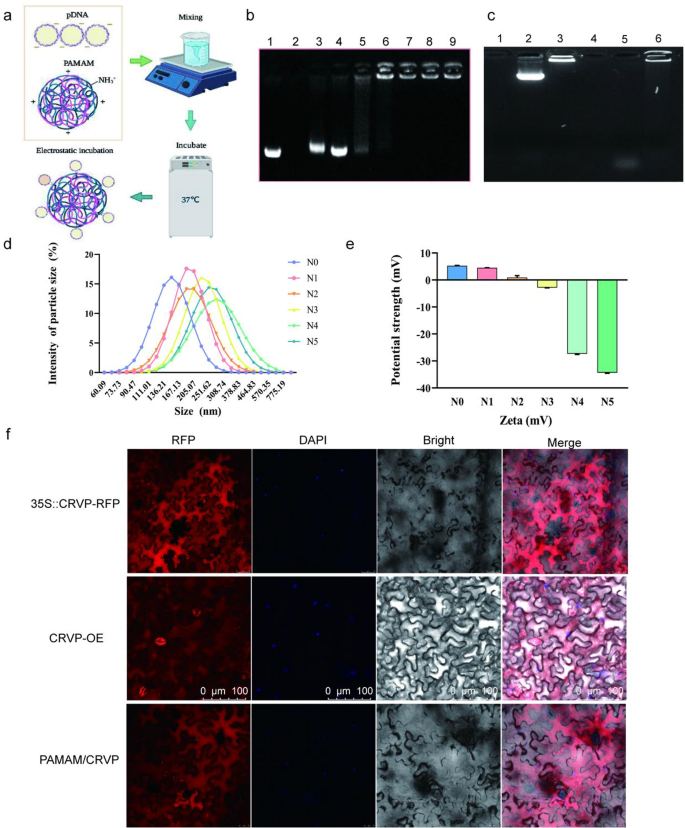
To realize environment friendly supply and expression of the resistant protein, we chosen PAMAM to organize plasmids-coated nanocomposites. The optimum enveloping situation was achieved by incubating at 37 ℃ for 30 min. PAMAM and expression plasmids will be mixed collectively to type nanocomposites beneath the motion of electrostatic binding power (Fig. 7a). Agarose gel electrophoresis was used to detect the binding capability of PAMAM to plasmids beneath completely different N/P ratios. When the N/P ratios have been 1:5 and 1:3, gel electrophoresis confirmed consultant bands with mobility much like the Mock-35S::CRVP plasmid. When the N/P ratio was 1:2, the 35S::CRVP plasmid could possibly be fully coated by PAMAM nanocarriers. General, the outcomes indicated that on the N/P ratio of 1:2, PAMAM exhibited the utmost loading restrict (Fig. 7b). The TEM and SEM photographs confirmed that PAMAM@CRVP complexes have been commonly dispersed and spherical and had the core dimension from 25 to 100 nm (Extra file 1: Fig. S6a, b). The outcomes of DNase remedy confirmed that the plasmids have been fully degraded after DNase remedy (37 °C, 10 min), whereas the PAMAM@pDNA nanocomposites have been solely partially degraded after DNase remedy in contrast with the management group, indicating that PAMAM might shield DNA from degradation by DNase (Fig. 7c).
Preparation and characterization of PAMAM@CRVP nanocomposites. a The method of getting ready PAMAM@CRVP nanocomposites. b The utmost plasmid loading capability of PAMAM. Lane 1: mock 35S::CRVP plasmids; Lane 2: mock PAMAM; and Lanes 3–9: PAMAM@CRVP nanocomposites with N/P ratios of 1:5, 1:3, 1:2, 1:1, 2:1, 3:1, and 5:1. c DNase remedy to detect the soundness of PAMAM@DNA. Lane 1: PAMAM; Lane 2: plasmids; Lane 3: PAMAM@DNA; Lane 4: PAMAM + DNase; Lane 5: plasmids + DNase; Lane 6: PAMAM@DNA+DNase. d The particle dimension of PAMAM@CRVP nanocomposites with completely different N/P ratios. N0: mock PAMAM; N1-5: nanocomposites with N/P ratios of 1:3, 1:2, 1:1, 2:1, and three:1, respectively. e The zeta potentials of PAMAM@CRVP nanocomposites with completely different N/P ratios. N0: mock PAMAM; N1-5: nanocomposites with N/P ratios of 1:3, 1:2, 1:1, 2:1, and three:1, respectively. f Laser confocal statement of NbCRVP expression in 35S::CRVP-RFP A. tumefaciens-infiltrated, CRVP-OE and PAMAM@CRVP nanocomposites handled vegetation.
The diameter of the nanocomposites was additional examined utilizing dynamic gentle scattering (DLS). PAMAM@CRVP nanocomposites at N/P ratios of 1:3, 1:2, 1:1, 2:1, and three:1 in addition to mock PAMAM had a mean dimension of 196.8 nm, 227.7 nm, 201.8 nm, 263.7 nm, 267.4 nm and 135.6 nm (Fig. 7d) and particle dispersion indices of 0.184, 0.442, 0.140, 0.195, 0.436 and 0.608, respectively (Extra file 2: Desk S2). The zeta potential worth of the PAMAM@CRVP nanocomposites was additionally examined in additional element utilizing a potentiometric analyzer. The typical zeta potentials of mock PAMAM and PAMAM@CRVP nanocomposites at N/P ratios of 1:3, 1:2, 1:1, 2:1, and three:1 have been 5.927 mV, 4.857 mV, 0.926 mV, 2.835 mV, -26.067 mV, and -33.667 mV, respectively (Fig. 7e), exhibiting a lower pattern upon incubation with negatively charged DNAs, and confirming that DNAs have been successfully combinated with PAMAM. Laser confocal microscopy was used to extra intuitively detect the expression of NbCRVP in vegetation handled with 35S::CRVP-RFP A. tumefaciens, CRVP-OE, and PAMAM@CRVP nanocomposites. The crimson fluorescence sign of NbCRVP-RFP fusion proteins was noticed in all three remedy teams, indicating that the PAMAM@CRVP nanocomplexs have been efficiently delivered into the plant cells (Fig. 7f). Protoplasts ready after infiltration of PAMAM@CRVP:CalB nanocomposites in Solanum lycopersicum and Capsicum annuum additionally confirmed the expression of NbCRVP and NbCalB (Extra file 1: Fig S7). Steady monitoring discovered that PAMAM-mediated gene expression was transient. On the ninth day after the PAMAM@CRVP:CalB nanocomplex infiltrated the plant leaves, the CRVP-RFP and CalB-GFP fluorescence alerts have been now not detectable within the cells (Extra file 1: Fig S8a). Nonetheless, the in vitro shelf life of those PAMAM@pDNA nanocomplexes might attain 28 days (Extra file 1: Fig S8b).
Optimization of the PAMAM@NbCRVP:NbCalB ratio for one of the best antiviral results
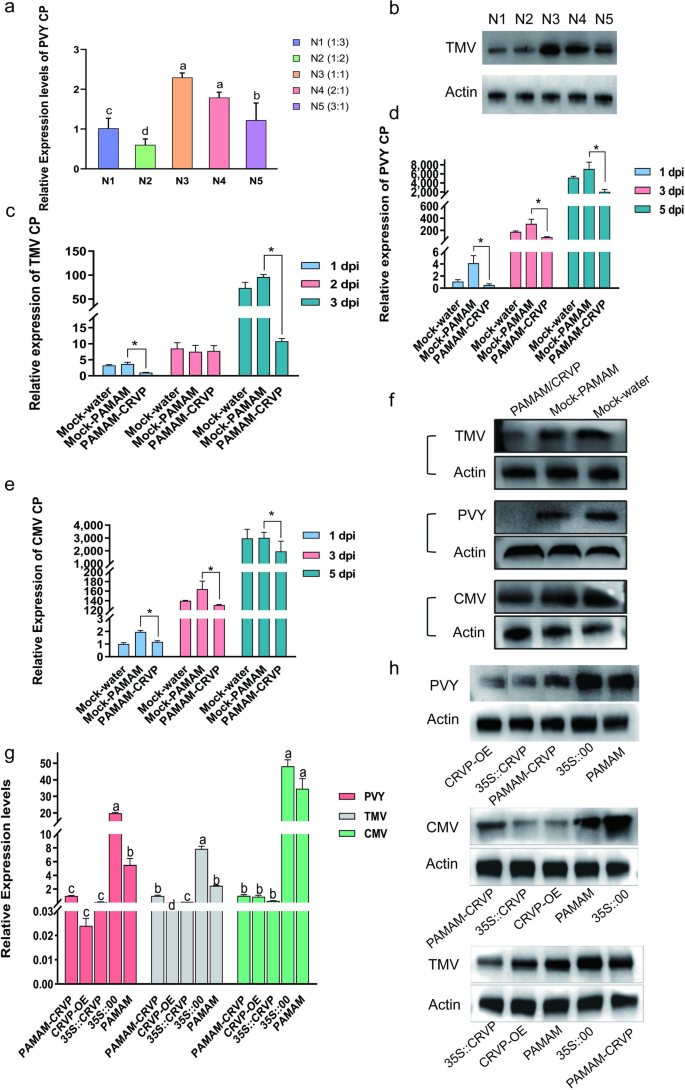
To find out the optimum ratio of PAMAM nanocarriers to CRVP plasmids, we examined their results on PVY an infection utilizing RT-qPCR on the N/P ratios of 1:3, 1:2, 1:1, 2:1, and three:1. It was discovered that PAMAM@CRVP had one of the best inhibitory impact on the N/P ratio of 1:2 (Fig. 8a), which was additional verified utilizing WB (Fig. 8b). Due to this fact, the N/P ratio of 1:2 was chosen because the optimum ratio for subsequent preparation of nanocomposites.
Antiviral results of PAMAM@CRVP nanocomposites. a The inhibitory results of PAMAM@CRVP nanocomposites with completely different N/P ratios on PVY an infection, as detected utilizing RT-qPCR. b The inhibitory results of PAMAM@CRVP nanocomposites with completely different N/P ratios on PVY an infection, as measured by WB. c The inhibitory impact of PAMAM@CRVP nanocomposites on TMV an infection at 1, 2, and three dpi, as detected by RT-qPCR. d The inhibitory impact of PAMAM@CRVP nanocomposites on PVY an infection at 1, 3, and 5 dpi, as detected by RT-qPCR. e The inhibitory impact of PAMAM@CRVP nanocomposites on CMV an infection at 1, 3, and 5 dpi, as detected by RT-qPCR. f The inhibitory impact of PAMAM@CRVP nanocomposites on PVY CMV and TMV infections, as detected by WB. g The inhibitory impact of PAMAM@CRVP nanocomposites on PVY CMV and TMV infections evaluating with CRVP-OE, 35S::CRVP, and 35S::00, PAMAM, as detected by RT-qPCR. h The inhibitory impact of PAMAM@CRVP nanocomposites on PVY CMV and TMV infections, evaluating with CRVP-OE, 35S::CRVP, 35S::00, and PAMAM, as measured by WB. Knowledge proven in a and g have been analyzed utilizing Duncan’s a number of vary exams within the ANOVA program of SPSS, with completely different letters indicating vital variations amongst completely different remedy teams (P < 0.05). Knowledge proven in c, d and e have been analyzed utilizing the impartial pattern t take a look at, with * indicating vital variations between two remedy teams (P < 0.05).
To confirm the anti-disease impact of PAMAM@CRVP nanocomposites, the PAMAM@CRVP nanocomposites have been examined towards TMV, PVY, and CMV an infection. RT-qPCR information confirmed that (1) TMV CP mRNA stage was decreased by 73% and 89% at 1 and three dpi, respectively, within the PAMAM@CRVP group in contrast with the management mock-PAMAM group (Fig. 8c); (2) PVY CP mRNA stage was decreased by 88%, 72%, and 71% at 1, 3, and 5 dpi, respectively, in contrast with the management mock-PAMAM group (Fig. 8d); and (3) CMV CP mRNA stage was decreased by 40%, 20%, and 35% at 1, 3, and 5 dpi, respectively, in contrast with the management mock-PAMAM group (Fig. 8e). WB outcomes additionally revealed decreased ranges of TMV CP at 3 dpi, PVY CP at 3 dpi, and CMV CP at 5 dpi, in line with the outcomes at mRNA ranges (Fig. 8f). These outcomes indicated that PAMAM@CRVP nanocomposites efficiently delivered the disease-resistant protein DNA and exerted virus-resistant impact. Moreover, A. tumefaciens-mediated remedy of vegetation with 35S::CRVP, and PAMAM@CRVP nanocomposites inhibited TMV, PVY, and CMV an infection, similar to the inhibitory impact with transgenic vegetation (Fig. 8g, h). These information additional confirmed that our examine offered a brand new supply platform for antiviral proteins.
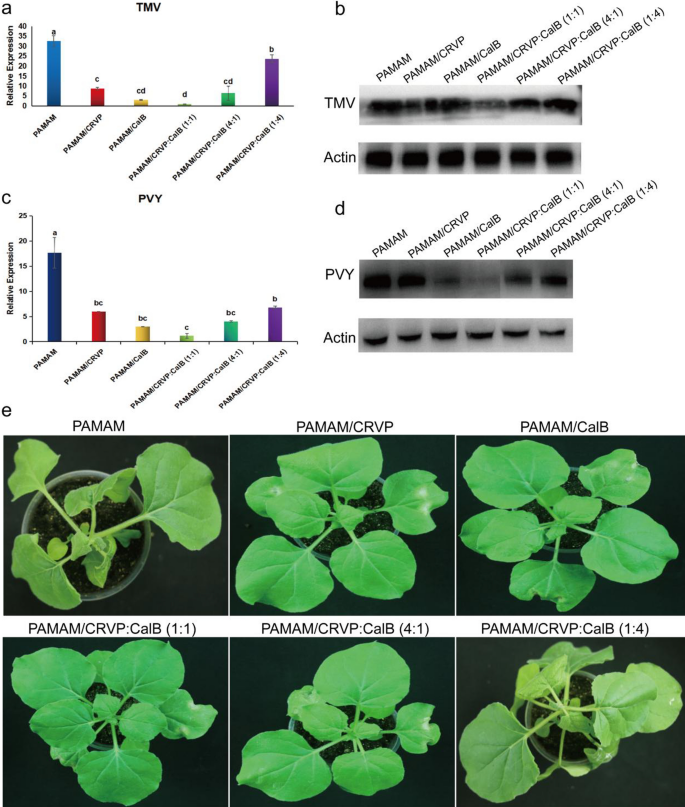
To make clear the optimum ratio of NbCRVP and NbCalB and their synergistic anti-disease impact, we arrange a number of therapies, together with mock-PAMAM, PAMAM@CRVP, PAMAM/CalB, PAMAM@CRVP:CalB (1:1), PAMAM@CRVP:CalB (4:1) and PAMAM@CRVP:CalB (1:4). qRT-PCR outcomes confirmed that PAMAM@CRVP, PAMAM/CalB, PAMAM@CRVP:CalB (1:1), PAMAM@CRVP:CalB (4:1), and PAMAM@CRVP:CalB (1:4) therapies downregulated the expression of TMV CP mRNA at 4 dpi by 73%, 90%, 97%, 80%, and 27%, respectively, with PAMAM@CRVP:CalB (1:1) exhibiting one of the best resistance impact (Fig. 9a). WB outcomes additional verified that TMV CP protein content material was the bottom within the PAMAM@CRVP:CalB (1:1) group (Fig. 9b). Furthermore, The PAMAM management group confirmed apparent signs of wilting and necrosis, whereas the PAMAM@CRVP:CalB (1:1) remedy group displayed no apparent virus an infection signs (Fig. 9e). We additionally examined the results of the above six remedy teams on PVY an infection. PVY CP mRNA (Fig. 9c) and protein (Fig. 9d) ranges have been the bottom within the PAMAM@CRVP:CalB (1:1) group, indicating the interplay between NbCRVP and NbCalB had a synergistic anti-disease impact, and NbCRVP:NbCalB on the ratio of 1:1 had one of the best anti-disease impact.
Interplay between NbCRVP and NbCalB has a synergistic anti-disease impact. a Impact of PAMAM nanocomposites with completely different NbCRVP:NbCalB ratios on TMV an infection at 4 dpi, as detected by RT-qPCR. b Results of PAMAM nanocomposites with completely different NbCRVP:NbCalB ratios on TMV an infection at 4 dpi, as detected by WB. c Impact of PAMAM nanocomposites with completely different NbCRVP:NbCalB ratios on PVY an infection at 4 dpi, as detected by RT-qPCR. d Results of PAMAM nanocomposites with completely different NbCRVP:NbCalB ratios on PVY an infection at 4 dpi, as detected by WB. e Impact of PAMAM nanocomposites with completely different NbCRVP:NbCalB ratios on the signs of TMV an infection at 4 dpi. The info proven in a and c have been analyzed by Duncan’s a number of vary exams within the ANOVA program of SPSS, with completely different letters indicating vital variations amongst completely different remedy teams (P < 0.05).