| Aug 01, 2023 |
|
(Nanowerk Information) A staff of scientists led by the College of Oxford have achieved a major breakthrough in detecting modifications on protein buildings. The tactic, printed in Nature Nanotechnology (“Enzyme-less nanopore detection of post-translational modifications inside lengthy polypeptides”), employs revolutionary nanopore expertise to establish structural variations on the single-molecule degree, even deep inside lengthy protein chains.
|
|
Human cells include roughly 20,000 protein-encoding genes. Nonetheless, the precise variety of proteins noticed in cells is way higher, with over 1,000,000 totally different buildings identified. These variants are generated by a course of referred to as post-translational modification (PTM), which happens after a protein has been transcribed from DNA. PTM introduces structural adjustments such because the addition of chemical teams or carbohydrate chains to the person amino acids that make up proteins. This leads to a whole lot of potential variations for a similar protein chain.
|
|
These variants play pivotal roles in biology, by enabling exact regulation of advanced organic processes inside particular person cells. Mapping this variation would uncover a wealth of useful data that would revolutionise our understanding of mobile features. However thus far, the power to provide complete protein inventories has remained an elusive purpose.
|
|
To beat this, a staff led by researchers on the College of Oxford’s Division of Chemistry has efficiently developed a technique for protein evaluation based mostly on nanopore DNA/RNA sequencing expertise. On this method, a directional circulation of water captures and unfolds 3D proteins into linear chains which can be fed by tiny pores, simply broad sufficient for a single amino acid molecule to move by. Structural variations are recognized by measuring adjustments in {an electrical} present utilized throughout the nanopore. Totally different molecules trigger totally different disruptions within the present, giving them a novel signature.
|
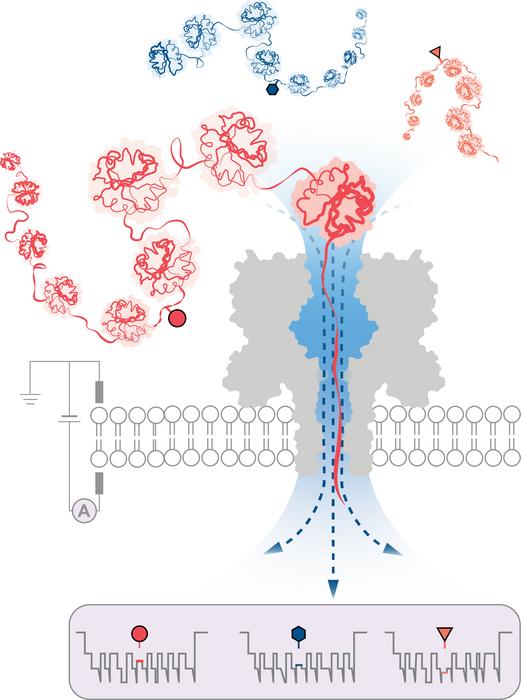 |
| An engineered protein nanopore directed a water flux sturdy sufficient to seize, unfold and translocate proteins exceeding 1200 amino acids in size. Modulation {of electrical} present throughout protein translocation by the nanopore detected post-translational modifications deep throughout the proteins (proven as circle, triangle, and hexagon). (Picture: Wei-Hsuan Lan and Yujia Qing)
|
|
The staff efficiently demonstrated the strategy’s effectiveness in detecting three totally different PTM modifications (phosphorylation, glutathionylation, and glycosylation) on the single-molecule degree for protein chains over 1,200 residues lengthy. These included modifications deep throughout the protein’s sequence. Importantly, the strategy doesn’t require the usage of labels, enzymes or extra reagents.
|
|
In keeping with the analysis staff, the brand new protein characterisation technique might be readily built-in into current transportable nanopore sequencing units to allow researchers to quickly construct protein inventories of single cells and tissues. This might facilitate point-of-care diagnostics, enabling the customized detection of particular protein variants related to illnesses together with most cancers and neurodegenerative issues.
|
|
Professor Yujia Qing (Division of Chemistry, College of Oxford), contributing creator for the research, mentioned: ‘This easy but highly effective technique opens up quite a few potentialities. Initially, it permits for the examination of particular person proteins, similar to these concerned in particular illnesses. In the long run, the strategy holds the potential to create prolonged inventories of protein variants inside cells, unlocking deeper insights into mobile processes and illness mechanisms.’
|
|
Professor Hagan Bayley (Division of Chemistry, College of Oxford), contributing creator and co-founder of Oxford Nanopore Applied sciences, added: ‘The flexibility to pinpoint and establish post-translational modifications and different protein variations on the single-molecule degree holds immense promise for advancing our understanding of mobile features and molecular interactions. It might additionally open new avenues for personalised medication, diagnostics, and therapeutic interventions.’
|