
Mapping the wiring and firing exercise of the human mind is key to deciphering how we predict — how we sense the world, be taught, resolve, keep in mind, and create — in addition to what points can come up in mind illness or dysfunction. Latest efforts have delivered publicly obtainable mind maps (high-resolution 3D mapping of mind cells and their connectivities) at unprecedented high quality and scale, akin to H01, a 1.4 petabyte nanometer-scale digital reconstruction of a pattern of human mind tissue from Harvard / Google, and the cubic millimeter mouse cortex dataset from our colleagues on the MICrONS consortium.
To interpret mind maps at this scale requires a number of layers of research, together with the identification of synaptic connections, mobile subcompartments, and cell sorts. Machine studying and pc imaginative and prescient know-how have performed a central function in enabling these analyses, however deploying such programs remains to be a laborious course of, requiring hours of guide floor reality labeling by professional annotators and vital computational assets. Furthermore, some necessary duties, akin to figuring out the cell kind from solely a small fragment of axon or dendrite, could be difficult even for human specialists, and haven’t but been successfully automated.
At the moment, in “Multi-Layered Maps of Neuropil with Segmentation-Guided Contrastive Studying”, we’re asserting Segmentation-Guided Contrastive Studying of Representations (SegCLR), a way for coaching wealthy, generic representations of mobile morphology (the cell’s form) and ultrastructure (the cell’s inside construction) with out laborious guide effort. SegCLR produces compact vector representations (i.e., embeddings) which are relevant throughout numerous downstream duties (e.g., native classification of mobile subcompartments, unsupervised clustering), and are even capable of determine cell sorts from solely small fragments of a cell. We educated SegCLR on each the H01 human cortex dataset and the MICrONS mouse cortex dataset, and we’re releasing the ensuing embedding vectors, about 8 billion in whole, for researchers to discover.
| From mind cells segmented out of a 3D block of tissue, SegCLR embeddings seize mobile morphology and ultrastructure and can be utilized to tell apart mobile subcompartments (e.g., dendritic backbone versus dendrite shaft) or cell sorts (e.g., pyramidal versus microglia cell). |
Representing Mobile Morphology and Ultrastructure
SegCLR builds on current advances in self-supervised contrastive studying. We use a regular deep community structure to encode inputs comprising native 3D blocks of electron microscopy knowledge (about 4 micrometers on a aspect) into 64-dimensional embedding vectors. The community is educated by way of a contrastive loss to map semantically associated inputs to related coordinates within the embedding area. That is near the in style SimCLR setup, besides that we additionally require an occasion segmentation of the amount (tracing out particular person cells and cell fragments), which we use in two necessary methods.
First, the enter 3D electron microscopy knowledge are explicitly masked by the segmentation, forcing the community to focus solely on the central cell inside every block. Second, we leverage the segmentation to routinely outline which inputs are semantically associated: constructive pairs for the contrastive loss are drawn from close by places on the identical segmented cell and educated to have related representations, whereas inputs drawn from totally different cells are educated to have dissimilar representations. Importantly, publicly obtainable automated segmentations of the human and mouse datasets had been sufficiently correct to coach SegCLR with out requiring laborious evaluation and correction by human specialists.
Decreasing Annotation Coaching Necessities by Three Orders of Magnitude
SegCLR embeddings can be utilized in numerous downstream settings, whether or not supervised (e.g., coaching classifiers) or unsupervised (e.g., clustering or content-based picture retrieval). Within the supervised setting, embeddings simplify the coaching of classifiers, and may significantly cut back floor reality labeling necessities. For instance, we discovered that for figuring out mobile subcompartments (axon, dendrite, soma, and so on.) a easy linear classifier educated on prime of SegCLR embeddings outperformed a completely supervised deep community educated on the identical activity, whereas utilizing solely about one thousand labeled examples as an alternative of tens of millions.
 |
| We assessed the classification efficiency for axon, dendrite, soma, and astrocyte subcompartments within the human cortex dataset by way of imply F1-Rating, whereas various the variety of coaching examples used. Linear classifiers educated on prime of SegCLR embeddings matched or exceeded the efficiency of a completely supervised deep classifier (horizontal line), whereas utilizing a fraction of the coaching knowledge. |
Distinguishing Cell Sorts, Even from Small Fragments
Distinguishing totally different cell sorts is a vital step in direction of understanding how mind circuits develop and performance in well being and illness. Human specialists can be taught to determine some cortical cell sorts primarily based on morphological options, however guide cell typing is laborious and ambiguous circumstances are frequent. Cell typing additionally turns into tougher when solely small fragments of cells can be found, which is frequent for a lot of cells in present connectomic reconstructions.
 |
| Human specialists manually labeled cell sorts for a small variety of proofread cells in every dataset. Within the mouse cortex dataset, specialists labeled six neuron sorts (prime) and 4 glia sorts (not proven). Within the human cortex dataset, specialists labeled two neuron sorts (not proven) and 4 glia sorts (backside). (Rows to not scale with one another.) |
We discovered that SegCLR precisely infers human and mouse cell sorts, even for small fragments. Previous to classification, we collected and averaged embeddings inside every cell over a set aggregation distance, outlined because the radius from a central level. We discovered that human cortical cell sorts could be recognized with excessive accuracy for aggregation radii as small as 10 micrometers, even for sorts that specialists discover tough to tell apart, akin to microglia (MGC) versus oligodendrocyte precursor cells (OPC).
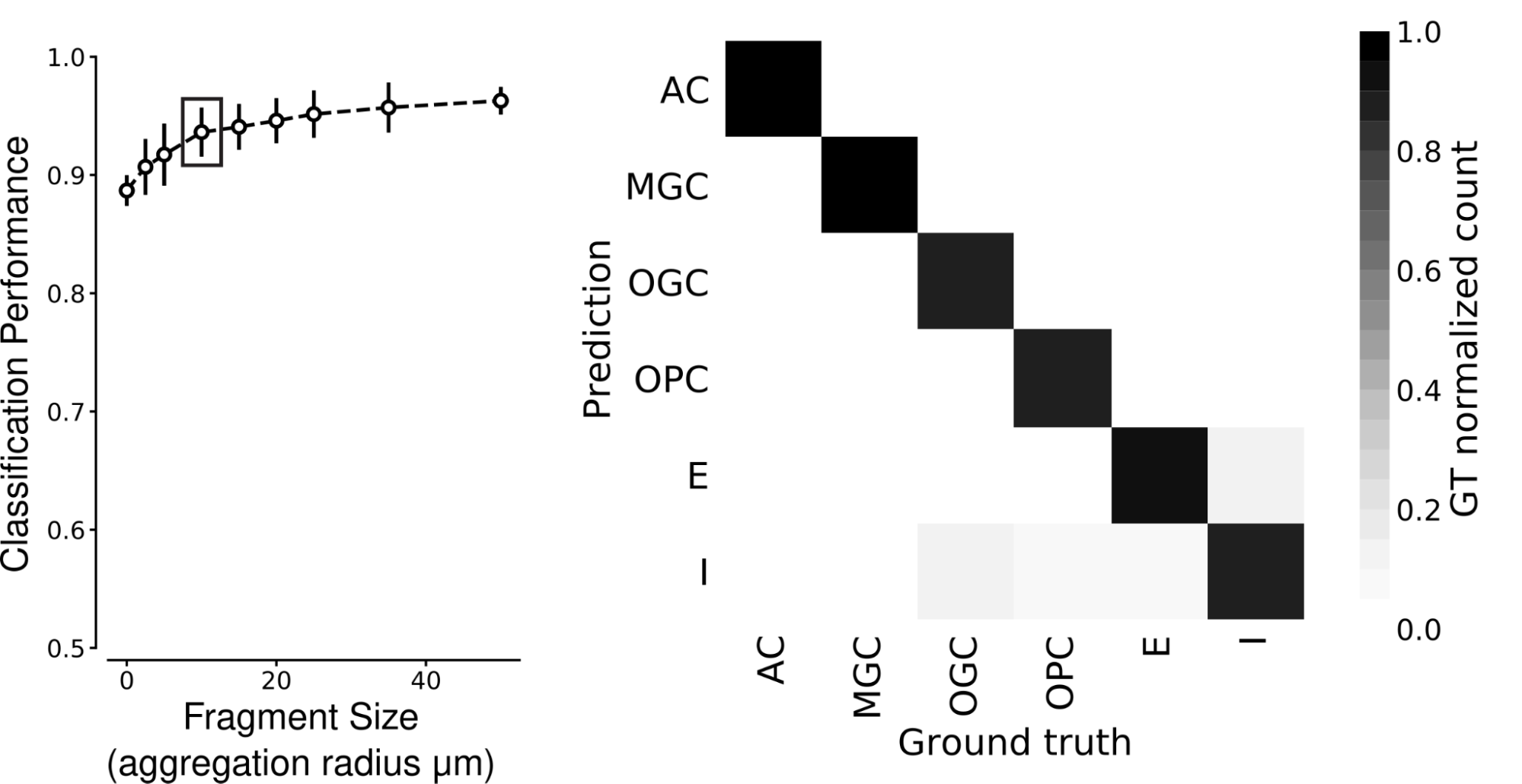 |
| SegCLR can classify cell sorts, even from small fragments. Left: Classification efficiency over six human cortex cell sorts for shallow ResNet fashions educated on SegCLR embeddings for various sized cell fragments. Aggregation radius zero corresponds to very small fragments with solely a single embedding. Cell kind efficiency reaches excessive accuracy (0.938 imply F1-Rating) for fragments with aggregation radii of solely 10 micrometers (boxed level). Proper: Class-wise confusion matrix at 10 micrometers aggregation radius. Darker shading alongside the diagonal signifies that predicted cell sorts agree with professional labels most often. AC: astrocyte; MGC: microglia cell; OGC: oligodendrocyte cell; OPC: oligodendrocyte precursor cell; E: excitatory neuron; I: inhibitory neuron. |
Within the mouse cortex, ten cell sorts could possibly be distinguished with excessive accuracy at aggregation radii of 25 micrometers.
 |
| Left: Classification efficiency over the ten mouse cortex cell sorts reaches 0.832 imply F1-Rating for fragments with aggregation radius 25 micrometers (boxed level). Proper: The category-wise confusion matrix at 25 micrometers aggregation radius. Bins point out broad teams (glia, excitatory neurons, and inhibitory interneurons). P: pyramidal cell; THLC: thalamocortical axon; BC: basket cell; BPC: bipolar cell; MC: Martinotti cell; NGC: neurogliaform cell. |
In extra cell kind purposes, we used unsupervised clustering of SegCLR embeddings to disclose additional neuronal subtypes, and demonstrated how uncertainty estimation can be utilized to prohibit classification to excessive confidence subsets of the dataset, e.g., when just a few cell sorts have professional labels.
Revealing Patterns of Mind Connectivity
Lastly, we confirmed how SegCLR can be utilized for automated evaluation of mind connectivity by cell typing the synaptic companions of reconstructed cells all through the mouse cortex dataset. Understanding the connectivity patterns between particular cell sorts is key to decoding large-scale connectomic reconstructions of mind wiring, however this usually requires guide tracing to determine accomplice cell sorts. Utilizing SegCLR, we replicated mind connectivity findings that beforehand relied on intensive guide tracing, whereas extending their scale when it comes to the variety of synapses, cell sorts, and mind areas analyzed. (See the paper for additional particulars.)
What’s Subsequent?
SegCLR captures wealthy mobile options and may significantly simplify downstream analyses in comparison with working straight with uncooked picture and segmentation knowledge. We’re excited to see what the group can uncover utilizing the ~8 billion embeddings we’re releasing for the human and mouse cortical datasets (instance entry code; browsable human and mouse views in Neuroglancer). By lowering complicated microscopy knowledge to wealthy and compact embedding representations, SegCLR opens many novel avenues for organic perception, and will function a hyperlink to complementary modalities for high-dimensional characterization on the mobile and subcellular ranges, akin to spatially-resolved transcriptomics.




