Introduction
On this put up we’ll describe methods to use smartphone accelerometer and gyroscope information to foretell the bodily actions of the people carrying the telephones. The information used on this put up comes from the Smartphone-Based mostly Recognition of Human Actions and Postural Transitions Knowledge Set distributed by the College of California, Irvine. Thirty people have been tasked with performing varied fundamental actions with an connected smartphone recording motion utilizing an accelerometer and gyroscope.
Earlier than we start, let’s load the varied libraries that we’ll use within the evaluation:
library(keras) # Neural Networks
library(tidyverse) # Knowledge cleansing / Visualization
library(knitr) # Desk printing
library(rmarkdown) # Misc. output utilities
library(ggridges) # VisualizationActions dataset
The information used on this put up come from the Smartphone-Based mostly Recognition of Human Actions and Postural Transitions Knowledge Set(Reyes-Ortiz et al. 2016) distributed by the College of California, Irvine.
When downloaded from the hyperlink above, the information incorporates two totally different ‘components.’ One which has been pre-processed utilizing varied characteristic extraction strategies resembling fast-fourier rework, and one other RawData part that merely provides the uncooked X,Y,Z instructions of an accelerometer and gyroscope. None of the usual noise filtering or characteristic extraction utilized in accelerometer information has been utilized. That is the information set we are going to use.
The motivation for working with the uncooked information on this put up is to assist the transition of the code/ideas to time sequence information in much less well-characterized domains. Whereas a extra correct mannequin may very well be made by using the filtered/cleaned information offered, the filtering and transformation can differ enormously from process to process; requiring plenty of guide effort and area information. One of many stunning issues about deep studying is the characteristic extraction is realized from the information, not outdoors information.
Exercise labels
The information has integer encodings for the actions which, whereas not necessary to the mannequin itself, are useful to be used to see. Let’s load them first.
activityLabels <- learn.desk("information/activity_labels.txt",
col.names = c("quantity", "label"))
activityLabels %>% kable(align = c("c", "l"))| 1 | WALKING |
| 2 | WALKING_UPSTAIRS |
| 3 | WALKING_DOWNSTAIRS |
| 4 | SITTING |
| 5 | STANDING |
| 6 | LAYING |
| 7 | STAND_TO_SIT |
| 8 | SIT_TO_STAND |
| 9 | SIT_TO_LIE |
| 10 | LIE_TO_SIT |
| 11 | STAND_TO_LIE |
| 12 | LIE_TO_STAND |
Subsequent, we load within the labels key for the RawData. This file is a listing of all the observations, or particular person exercise recordings, contained within the information set. The important thing for the columns is taken from the information README.txt.
Column 1: experiment quantity ID,
Column 2: person quantity ID,
Column 3: exercise quantity ID
Column 4: Label begin level
Column 5: Label finish level The beginning and finish factors are in variety of sign log samples (recorded at 50hz).
Let’s check out the primary 50 rows:
labels <- learn.desk(
"information/RawData/labels.txt",
col.names = c("experiment", "userId", "exercise", "startPos", "endPos")
)
labels %>%
head(50) %>%
paged_table()File names
Subsequent, let’s have a look at the precise recordsdata of the person information offered to us in RawData/
dataFiles <- checklist.recordsdata("information/RawData")
dataFiles %>% head()
[1] "acc_exp01_user01.txt" "acc_exp02_user01.txt"
[3] "acc_exp03_user02.txt" "acc_exp04_user02.txt"
[5] "acc_exp05_user03.txt" "acc_exp06_user03.txt"There’s a three-part file naming scheme. The primary half is the kind of information the file incorporates: both acc for accelerometer or gyro for gyroscope. Subsequent is the experiment quantity, and final is the person Id for the recording. Let’s load these right into a dataframe for ease of use later.
fileInfo <- data_frame(
filePath = dataFiles
) %>%
filter(filePath != "labels.txt") %>%
separate(filePath, sep = '_',
into = c("sort", "experiment", "userId"),
take away = FALSE) %>%
mutate(
experiment = str_remove(experiment, "exp"),
userId = str_remove_all(userId, "person|.txt")
) %>%
unfold(sort, filePath)
fileInfo %>% head() %>% kable()| 01 | 01 | acc_exp01_user01.txt | gyro_exp01_user01.txt |
| 02 | 01 | acc_exp02_user01.txt | gyro_exp02_user01.txt |
| 03 | 02 | acc_exp03_user02.txt | gyro_exp03_user02.txt |
| 04 | 02 | acc_exp04_user02.txt | gyro_exp04_user02.txt |
| 05 | 03 | acc_exp05_user03.txt | gyro_exp05_user03.txt |
| 06 | 03 | acc_exp06_user03.txt | gyro_exp06_user03.txt |
Studying and gathering information
Earlier than we will do something with the information offered we have to get it right into a model-friendly format. This implies we need to have a listing of observations, their class (or exercise label), and the information similar to the recording.
To acquire this we are going to scan by way of every of the recording recordsdata current in dataFiles, lookup what observations are contained within the recording, extract these recordings and return every thing to a simple to mannequin with dataframe.
# Learn contents of single file to a dataframe with accelerometer and gyro information.
readInData <- perform(experiment, userId){
genFilePath = perform(sort) {
paste0("information/RawData/", sort, "_exp",experiment, "_user", userId, ".txt")
}
bind_cols(
learn.desk(genFilePath("acc"), col.names = c("a_x", "a_y", "a_z")),
learn.desk(genFilePath("gyro"), col.names = c("g_x", "g_y", "g_z"))
)
}
# Operate to learn a given file and get the observations contained alongside
# with their courses.
loadFileData <- perform(curExperiment, curUserId) {
# load sensor information from file into dataframe
allData <- readInData(curExperiment, curUserId)
extractObservation <- perform(startPos, endPos){
allData[startPos:endPos,]
}
# get statement areas on this file from labels dataframe
dataLabels <- labels %>%
filter(userId == as.integer(curUserId),
experiment == as.integer(curExperiment))
# extract observations as dataframes and save as a column in dataframe.
dataLabels %>%
mutate(
information = map2(startPos, endPos, extractObservation)
) %>%
choose(-startPos, -endPos)
}
# scan by way of all experiment and userId combos and collect information right into a dataframe.
allObservations <- map2_df(fileInfo$experiment, fileInfo$userId, loadFileData) %>%
right_join(activityLabels, by = c("exercise" = "quantity")) %>%
rename(activityName = label)
# cache work.
write_rds(allObservations, "allObservations.rds")
allObservations %>% dim()Exploring the information
Now that we now have all the information loaded together with the experiment, userId, and exercise labels, we will discover the information set.
Size of recordings
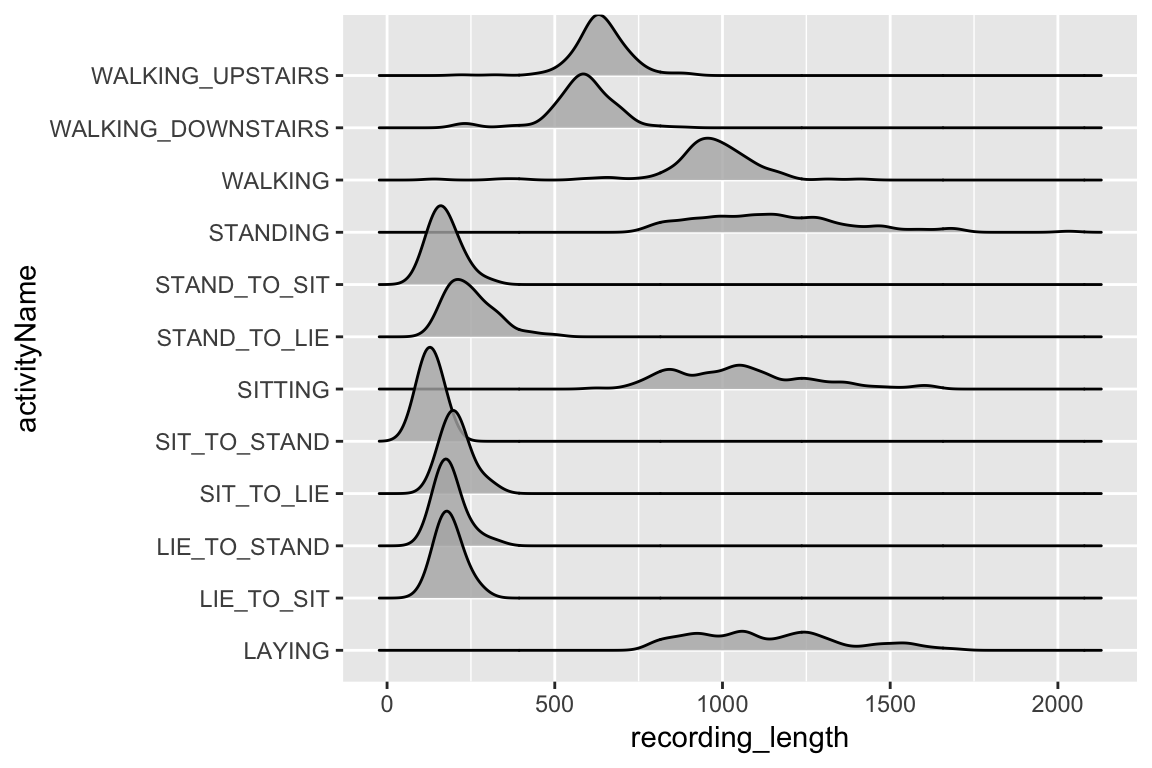
Let’s first have a look at the size of the recordings by exercise.
allObservations %>%
mutate(recording_length = map_int(information,nrow)) %>%
ggplot(aes(x = recording_length, y = activityName)) +
geom_density_ridges(alpha = 0.8)
The actual fact there may be such a distinction in size of recording between the totally different exercise sorts requires us to be a bit cautious with how we proceed. If we prepare the mannequin on each class directly we’re going to must pad all of the observations to the size of the longest, which would go away a big majority of the observations with an enormous proportion of their information being simply padding-zeros. Due to this, we are going to match our mannequin to simply the most important ‘group’ of observations size actions, these embody STAND_TO_SIT, STAND_TO_LIE, SIT_TO_STAND, SIT_TO_LIE, LIE_TO_STAND, and LIE_TO_SIT.
An attention-grabbing future course can be making an attempt to make use of one other structure resembling an RNN that may deal with variable size inputs and coaching it on all the information. Nevertheless, you’d run the danger of the mannequin studying merely that if the statement is lengthy it’s most definitely one of many 4 longest courses which might not generalize to a state of affairs the place you have been working this mannequin on a real-time-stream of information.
Filtering actions
Based mostly on our work from above, let’s subset the information to simply be of the actions of curiosity.
desiredActivities <- c(
"STAND_TO_SIT", "SIT_TO_STAND", "SIT_TO_LIE",
"LIE_TO_SIT", "STAND_TO_LIE", "LIE_TO_STAND"
)
filteredObservations <- allObservations %>%
filter(activityName %in% desiredActivities) %>%
mutate(observationId = 1:n())
filteredObservations %>% paged_table()So after our aggressive pruning of the information we can have a decent quantity of information left upon which our mannequin can study.
Coaching/testing break up
Earlier than we go any additional into exploring the information for our mannequin, in an try and be as honest as doable with our efficiency measures, we have to break up the information right into a prepare and check set. Since every person carried out all actions simply as soon as (apart from one who solely did 10 of the 12 actions) by splitting on userId we are going to be sure that our mannequin sees new folks solely after we check it.
# get all customers
userIds <- allObservations$userId %>% distinctive()
# randomly select 24 (80% of 30 people) for coaching
set.seed(42) # seed for reproducibility
trainIds <- pattern(userIds, dimension = 24)
# set the remainder of the customers to the testing set
testIds <- setdiff(userIds,trainIds)
# filter information.
trainData <- filteredObservations %>%
filter(userId %in% trainIds)
testData <- filteredObservations %>%
filter(userId %in% testIds)Visualizing actions
Now that we now have trimmed our information by eradicating actions and splitting off a check set, we will truly visualize the information for every class to see if there’s any instantly discernible form that our mannequin could possibly choose up on.
First let’s unpack our information from its dataframe of one-row-per-observation to a tidy model of all of the observations.
unpackedObs <- 1:nrow(trainData) %>%
map_df(perform(rowNum){
dataRow <- trainData[rowNum, ]
dataRow$information[[1]] %>%
mutate(
activityName = dataRow$activityName,
observationId = dataRow$observationId,
time = 1:n() )
}) %>%
collect(studying, worth, -time, -activityName, -observationId) %>%
separate(studying, into = c("sort", "course"), sep = "_") %>%
mutate(sort = ifelse(sort == "a", "acceleration", "gyro"))Now we now have an unpacked set of our observations, let’s visualize them!
unpackedObs %>%
ggplot(aes(x = time, y = worth, shade = course)) +
geom_line(alpha = 0.2) +
geom_smooth(se = FALSE, alpha = 0.7, dimension = 0.5) +
facet_grid(sort ~ activityName, scales = "free_y") +
theme_minimal() +
theme( axis.textual content.x = element_blank() )
So no less than within the accelerometer information patterns undoubtedly emerge. One would think about that the mannequin might have bother with the variations between LIE_TO_SIT and LIE_TO_STAND as they’ve an identical profile on common. The identical goes for SIT_TO_STAND and STAND_TO_SIT.
Preprocessing
Earlier than we will prepare the neural community, we have to take a few steps to preprocess the information.
Padding observations
First we are going to resolve what size to pad (and truncate) our sequences to by discovering what the 98th percentile size is. By not utilizing the very longest statement size this may assist us keep away from extra-long outlier recordings messing up the padding.
padSize <- trainData$information %>%
map_int(nrow) %>%
quantile(p = 0.98) %>%
ceiling()
padSize
98%
334 Now we merely have to convert our checklist of observations to matrices, then use the tremendous helpful pad_sequences() perform in Keras to pad all observations and switch them right into a 3D tensor for us.
convertToTensor <- . %>%
map(as.matrix) %>%
pad_sequences(maxlen = padSize)
trainObs <- trainData$information %>% convertToTensor()
testObs <- testData$information %>% convertToTensor()
dim(trainObs)
[1] 286 334 6Fantastic, we now have our information in a pleasant neural-network-friendly format of a 3D tensor with dimensions (<num obs>, <sequence size>, <channels>).
One-hot encoding
There’s one last item we have to do earlier than we will prepare our mannequin, and that’s flip our statement courses from integers into one-hot, or dummy encoded, vectors. Fortunately, once more Keras has provided us with a really useful perform to do exactly this.
oneHotClasses <- . %>%
{. - 7} %>% # convey integers all the way down to 0-6 from 7-12
to_categorical() # One-hot encode
trainY <- trainData$exercise %>% oneHotClasses()
testY <- testData$exercise %>% oneHotClasses()Modeling
Structure
Since we now have temporally dense time-series information we are going to make use of 1D convolutional layers. With temporally-dense information, an RNN has to study very lengthy dependencies in an effort to choose up on patterns, CNNs can merely stack a number of convolutional layers to construct sample representations of considerable size. Since we’re additionally merely in search of a single classification of exercise for every statement, we will simply use pooling to ‘summarize’ the CNNs view of the information right into a dense layer.
Along with stacking two layer_conv_1d() layers, we are going to use batch norm and dropout (the spatial variant(Tompson et al. 2014) on the convolutional layers and normal on the dense) to regularize the community.
input_shape <- dim(trainObs)[-1]
num_classes <- dim(trainY)[2]
filters <- 24 # variety of convolutional filters to study
kernel_size <- 8 # what number of time-steps every conv layer sees.
dense_size <- 48 # dimension of our penultimate dense layer.
# Initialize mannequin
mannequin <- keras_model_sequential()
mannequin %>%
layer_conv_1d(
filters = filters,
kernel_size = kernel_size,
input_shape = input_shape,
padding = "legitimate",
activation = "relu"
) %>%
layer_batch_normalization() %>%
layer_spatial_dropout_1d(0.15) %>%
layer_conv_1d(
filters = filters/2,
kernel_size = kernel_size,
activation = "relu",
) %>%
# Apply common pooling:
layer_global_average_pooling_1d() %>%
layer_batch_normalization() %>%
layer_dropout(0.2) %>%
layer_dense(
dense_size,
activation = "relu"
) %>%
layer_batch_normalization() %>%
layer_dropout(0.25) %>%
layer_dense(
num_classes,
activation = "softmax",
title = "dense_output"
)
abstract(mannequin)
______________________________________________________________________
Layer (sort) Output Form Param #
======================================================================
conv1d_1 (Conv1D) (None, 327, 24) 1176
______________________________________________________________________
batch_normalization_1 (BatchNo (None, 327, 24) 96
______________________________________________________________________
spatial_dropout1d_1 (SpatialDr (None, 327, 24) 0
______________________________________________________________________
conv1d_2 (Conv1D) (None, 320, 12) 2316
______________________________________________________________________
global_average_pooling1d_1 (Gl (None, 12) 0
______________________________________________________________________
batch_normalization_2 (BatchNo (None, 12) 48
______________________________________________________________________
dropout_1 (Dropout) (None, 12) 0
______________________________________________________________________
dense_1 (Dense) (None, 48) 624
______________________________________________________________________
batch_normalization_3 (BatchNo (None, 48) 192
______________________________________________________________________
dropout_2 (Dropout) (None, 48) 0
______________________________________________________________________
dense_output (Dense) (None, 6) 294
======================================================================
Complete params: 4,746
Trainable params: 4,578
Non-trainable params: 168
______________________________________________________________________Coaching
Now we will prepare the mannequin utilizing our check and coaching information. Word that we use callback_model_checkpoint() to make sure that we save solely the most effective variation of the mannequin (fascinating since in some unspecified time in the future in coaching the mannequin might start to overfit or in any other case cease enhancing).
# Compile mannequin
mannequin %>% compile(
loss = "categorical_crossentropy",
optimizer = "rmsprop",
metrics = "accuracy"
)
trainHistory <- mannequin %>%
match(
x = trainObs, y = trainY,
epochs = 350,
validation_data = checklist(testObs, testY),
callbacks = checklist(
callback_model_checkpoint("best_model.h5",
save_best_only = TRUE)
)
)
The mannequin is studying one thing! We get a decent 94.4% accuracy on the validation information, not unhealthy with six doable courses to select from. Let’s look into the validation efficiency somewhat deeper to see the place the mannequin is messing up.
Analysis
Now that we now have a skilled mannequin let’s examine the errors that it made on our testing information. We are able to load the most effective mannequin from coaching based mostly upon validation accuracy after which have a look at every statement, what the mannequin predicted, how excessive a chance it assigned, and the true exercise label.
# dataframe to get labels onto one-hot encoded prediction columns
oneHotToLabel <- activityLabels %>%
mutate(quantity = quantity - 7) %>%
filter(quantity >= 0) %>%
mutate(class = paste0("V",quantity + 1)) %>%
choose(-number)
# Load our greatest mannequin checkpoint
bestModel <- load_model_hdf5("best_model.h5")
tidyPredictionProbs <- bestModel %>%
predict(testObs) %>%
as_data_frame() %>%
mutate(obs = 1:n()) %>%
collect(class, prob, -obs) %>%
right_join(oneHotToLabel, by = "class")
predictionPerformance <- tidyPredictionProbs %>%
group_by(obs) %>%
summarise(
highestProb = max(prob),
predicted = label[prob == highestProb]
) %>%
mutate(
fact = testData$activityName,
appropriate = fact == predicted
)
predictionPerformance %>% paged_table()First, let’s have a look at how ‘assured’ the mannequin was by if the prediction was appropriate or not.
predictionPerformance %>%
mutate(end result = ifelse(appropriate, 'Appropriate', 'Incorrect')) %>%
ggplot(aes(highestProb)) +
geom_histogram(binwidth = 0.01) +
geom_rug(alpha = 0.5) +
facet_grid(end result~.) +
ggtitle("Chances related to prediction by correctness")
Reassuringly it appears the mannequin was, on common, much less assured about its classifications for the inaccurate outcomes than the proper ones. (Though, the pattern dimension is simply too small to say something definitively.)
Let’s see what actions the mannequin had the toughest time with utilizing a confusion matrix.
predictionPerformance %>%
group_by(fact, predicted) %>%
summarise(depend = n()) %>%
mutate(good = fact == predicted) %>%
ggplot(aes(x = fact, y = predicted)) +
geom_point(aes(dimension = depend, shade = good)) +
geom_text(aes(label = depend),
hjust = 0, vjust = 0,
nudge_x = 0.1, nudge_y = 0.1) +
guides(shade = FALSE, dimension = FALSE) +
theme_minimal()
We see that, because the preliminary visualization advised, the mannequin had a little bit of bother with distinguishing between LIE_TO_SIT and LIE_TO_STAND courses, together with the SIT_TO_LIE and STAND_TO_LIE, which even have comparable visible profiles.
Future instructions
The obvious future course to take this evaluation can be to aim to make the mannequin extra common by working with extra of the provided exercise sorts. One other attention-grabbing course can be to not separate the recordings into distinct ‘observations’ however as an alternative hold them as one streaming set of information, very like an actual world deployment of a mannequin would work, and see how effectively a mannequin might classify streaming information and detect adjustments in exercise.
Gal, Yarin, and Zoubin Ghahramani. 2016. “Dropout as a Bayesian Approximation: Representing Mannequin Uncertainty in Deep Studying.” In Worldwide Convention on Machine Studying, 1050–9.
Graves, Alex. 2012. “Supervised Sequence Labelling.” In Supervised Sequence Labelling with Recurrent Neural Networks, 5–13. Springer.
Kononenko, Igor. 1989. “Bayesian Neural Networks.” Organic Cybernetics 61 (5). Springer: 361–70.
LeCun, Yann, Yoshua Bengio, and Geoffrey Hinton. 2015. “Deep Studying.” Nature 521 (7553). Nature Publishing Group: 436.
Reyes-Ortiz, Jorge-L, Luca Oneto, Albert Samà, Xavier Parra, and Davide Anguita. 2016. “Transition-Conscious Human Exercise Recognition Utilizing Smartphones.” Neurocomputing 171. Elsevier: 754–67.
Tompson, Jonathan, Ross Goroshin, Arjun Jain, Yann LeCun, and Christoph Bregler. 2014. “Environment friendly Object Localization Utilizing Convolutional Networks.” CoRR abs/1411.4280. http://arxiv.org/abs/1411.4280.