Cells and animals
H9C2 and HEK-293T cells have been bought from the Nationwide Assortment of Authenticated Cell Cultures, Chinese language Academy of Sciences (China); they have been cultured in DMEM (excessive glucose) medium (01-055-1A, Organic Industries, Israel) supplemented with 10% foetal bovine serum (04-010-1A, Organic Industries, Israel). The cells have been stored at 37 °C in 5% CO2 at atmospheric stress.
Male Sprague Dawley rats (250 ± 10 g, 8 weeks previous) have been bought from the Mannequin Animal Analysis Middle of Nantong College. The rats have been stored in a selected pathogen-free (SPF) setting at 22 ± 1 °C, a relative humidity of fifty ± 1%, and a daily 12 h day-night cycle. All animal experiments have been authorised by the Medical Ethics Committee of Nantong College and performed underneath the steering of the Laboratory of Nantong College. This research was authorised by the Ethics Committee of the Affiliated Hospital of Nantong College (No. S20200314-012).
In vitro MIRI cell mannequin and in vivo MIRI rat mannequin
For the in vitro experiments, H9C2 cells have been cultured in full medium (DMEM with 10% FBS) to 80% confluence. The cells have been then cultured in serum and sugar-free medium at 37 °C in a three-atmosphere incubator underneath hypoxic situations for six h, which consisted of 94% N2, 5% CO2, and 1% O2. Lastly, the cells have been cultured in full medium underneath common oxygen situations for 12 h to ascertain reoxidation. H9C2 cells that weren’t uncovered to hypoxic situations have been thought of the management group.
For the in vivo experiments, 8 week-old male Sprague Dawley rats have been used. The rats have been randomly assigned to the completely different teams. Animal care was performed in accordance with the Institutional Animal Care and Use Committee (IACUC) tips. Preparation of the rat myocardial ischaemia reperfusion mannequin: Rats have been administered isoflurane all through the surgical procedure for anaesthesia. They have been artificially ventilated by way of a tracheal intubation tube related to a VentStar Small Animal Ventilator (R415, RWD, China). The pores and skin of the rat’s chest was depilated and disinfected with iodine. The pores and skin was minimize alongside the longitudinal sternum, the subcutaneous tissues and muscle mass have been freed layer by layer with haemostatic forceps, the left third to 4th intercostal house was uncovered, and the darkish shadow of the pulsating coronary heart could possibly be clearly seen. The rib cage was separated by inserting elbow haemostatic forceps into the thoracic cavity alongside the left fringe of the sternum on the 3–4 rib house. The pericardium was minimize open after opening the thoracic cavity and the guts was totally uncovered with a chest opener. The left anterior descending department of the coronary artery was ligated with 6/0 medical sutures for 30 min to dam blood perfusion after which launched for reperfusion, and the thoracic cavity was sutured layer by layer.
Preparation of PMVs@PLGA-miRNA complexes
Preparation of platelet membrane vesicles (PMVs)
Blood from SD male rats was collected into an EDTA tube after which centrifuged at 200 × g for 20 min at room temperature (RT) to separate the pink and white blood cells. The supernatant, platelet-rich plasma (PRP) containing the platelets, was collected. Phosphate-buffered saline (PBS) with 5 mM prostaglandin E1 (PGE1) (GC41905, GlpBio, USA) was added to the purified PRP to maintain the platelets inactivated. The remoted platelets have been then pelleted by centrifugation at 1800 ×g for 20 min at RT. After eradicating the supernatant, the platelets have been resuspended in PBS containing 5 mM PGE1. The ready suspended platelets have been counted in a move cytometer, and the variety of platelets per tube (2.5–3.0) ×105 was standardized. To manufacture PMVs, pelleted platelets from SD male rat plasma have been repeatedly freeze‒thawed, centrifuged at 8000 ×g for 15 min, after which sonicated for two min in a shower sonicator (FS30D, Fisher Scientific, USA) [7]. The platelet membranes have been filtered by way of a sterile 0.2 μm filter to provide the PMVs. The PMVs have been aliquoted into 1 mL samples and positioned at − 80 °C for storage till use. The scale distribution and morphology of the PMVs have been examined utilizing transmission electron microscopy.
Preparation of PLGA and PLGA-miRNA complexes
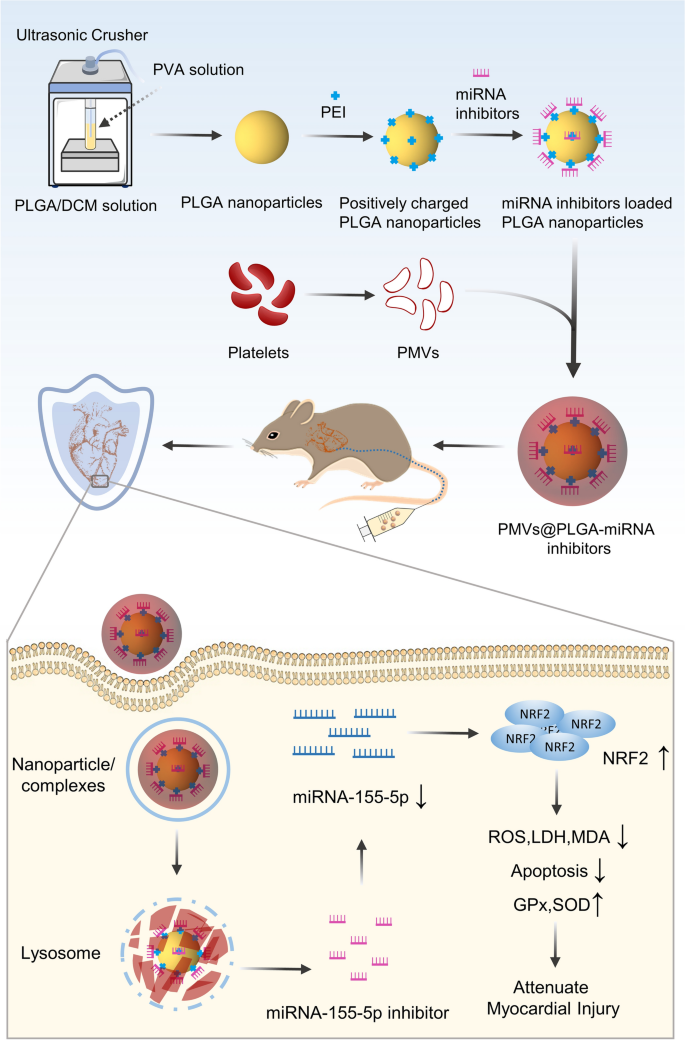
PLGA nanoparticles have been obtained by the double emulsion methodology as reported in a earlier research [30, 31]. The primary part of the nanoparticles is poly (D,L-lactide-co-glycolide) (PLGA, lactide:glycolide (65:35), Mw = 40–75 kDa) (P2066, Sigma‒Aldrich, USA). To load the miRNAs into the nanoparticles, the nanoparticles have been modified with branched polyethyleneimine (PEI, Mw = 25 kDa) (408727, Sigma‒Aldrich, USA) to make them positively charged so they might entice the negatively charged miRNAs. The miR-140-5p, miR-144-3p, miR-155-5p, and miR-340-5p mimic, inhibitor, NC mimic, NC inhibitor, and miR-155-5p antagomir have been bought from GenePharma (China). Briefly, the nanoparticle resolution was blended with PEI in deionized water, after which the blended resolution was added to the miRNA options, vortexed gently and incubated for 20 min at RT to formulate the PLGA-miRNA complexes.
Preparation of PMVs@PLGA and PMVs@PLGA-miRNA complexes
The PMVs vesicles have been fused with an equal quantity of PLGA or PLGA-miRNA complexes by ultrasound (5 min, 42 kHz, 100 W). The pattern was filtered 20 occasions utilizing a porous syringe filter with a membrane pore measurement of 200 nm and centrifuged (2500 rpm, 10 min) to take away extra PMVs to provide the PMVs@PLGA or PMVs@PLGA-miRNA complexes (Fig. 1).
Characterization of the PMVs@PLGA complexes in vitro
The characterization of the PMVs@PLGA complexes in vitro was much like a earlier research [29,30,31]. The scale and morphology of the PLGA-miRNA complexes have been characterised by transmission electron microscopy (TEM) (JEM-2100, JEOL, Japan). Briefly, the complexes have been coated with platinum after freeze-drying after which imaged underneath TEM. The common diameter and measurement distribution of the complexes have been decided by nanoparticle monitoring evaluation (NTA) (ZetaView, Particle Metrix, Germany). A gel retardation assay was used to judge the interplay between the PLGA nanoparticles and miRNAs at completely different N/M ratios (the ratio of moles of the amine teams of polyethyleneimine to the moles of the phosphate teams of RNA). The in vitro cumulative launch of miRNA from the PMVs@PLGA-miRNA complexes was measured. Briefly, complexes containing 12 μg miRNA inhibitors have been ready. The complexes have been equally divided into three tubes, and 50 μL PBS was added to every tube and incubated at 37 °C with light agitation. The supernatant was collected each two days by centrifugation at 15,000 rpm for five min and changed with recent PBS. The collected supernatant was used to detect siRNA concentrations with a Quant-iT RNA Assay Package (Q33140, Invitrogen, USA). Lastly, we calculated the discharge quantity and plotted the miRNA launch profile. A crude comparability of the proteins in platelets and the PMVs, PLGA and PMVs@PLGA complexes was carried out utilizing a Coomassie Blue Staining Package (P0017A, Beyotime, China). The loading weight of the proteins for every lane was 20 μg. Platelet membrane markers have been detected by western blotting (WB), and the primary markers have been CD31, CD41, CD42b, and CD61. The biocompatibility of PMVs, PLGA and PMVs@PLGA complexes with H9C2 cells was evaluated by CCK-8 assays in vitro. Thick smears have been made for commentary by fluorescence microscopy utilizing PKH67-labelled PMVs and rhodamine-labelled PLGA.
When utilizing move cytometry to detect the particle uptake effectivity, Dil-labelled PMVs and FITC-labelled PLGA have been used. The toxicity of the PMVs, PLGA and PMVs@PLGA complexes to cells was additionally detected by move cytometry of H9C2 apoptosis. As well as, TUNEL apoptosis, caspase 3 exercise, ROS, complete SOD exercise, MDA, GPx enzyme exercise and LDH cytotoxicity have been all used to evaluate their toxicity additional. A haemolytic assay was additionally used as a part of the protection test.
Reactive oxygen species (ROS) assay
The ROS assay was accomplished utilizing a Reactive Oxygen Species Assay Package (S0033 M, Beyotime, China). Handled H9C2 cells have been labelled utilizing a DCFH-DA probe (10 μmol/L), incubated for 20 min at 37 °C, washed with PBS to take away extra probe after which assayed utilizing move cytometry.
Complete superoxide dismutase (SOD) exercise assay
Complete SOD exercise was measured utilizing a Cu/Zn-SOD and Mn-SOD Assay Package with WST-8 (S0103, Beyotime, China). After getting ready the working resolution and treating the cells in line with the producer’s directions, the absorbance of every pattern was measured at 450 nm and the full SOD exercise was calculated as described within the handbook.
Malondialdehyde (MDA) assay
MDA was measured utilizing a Malondialdehyde (MDA) Content material Check Package (BC0020, Solarbio, China). After getting ready the working resolution and treating the cells in line with the producer’s directions, the absorbance of every pattern was measured at 532 nm and 600 nm and the MDA content material was calculated as described within the handbook.
Glutathione peroxidase (GPx) enzyme exercise assay
GPx enzyme exercise was measured utilizing a Complete Glutathione Peroxidase Assay Package with NADPH (S0058, Beyotime, China). After getting ready the working resolution and treating the cells in line with the producer’s directions, the absorbance of every pattern was measured at 340 nm and the GPx enzyme exercise was calculated as described within the handbook.
Lactate dehydrogenase (LDH) cytotoxicity assay
LDH cytotoxicity was measured utilizing a LDH Cytotoxicity Assay Package (C0017, Beyotime, China). After getting ready the working resolution and treating the cells in line with the producer’s directions, the absorbance of every pattern was measured at 490 nm and 600 nm and the LDH cytotoxicity was calculated as described within the handbook.
Haemolytic assay
Blood was collected from wholesome SD rats and centrifuged at 400 × g for 15 min to isolate the pink blood cells. The cells have been then washed 3 occasions with PBS, and PMVs, PLGA or PMVs@PLGA complexes suspended in PBS have been blended with the erythrocytes at a quantity ratio of 6:4 and incubated for 3 h at 37 °C (a PBS-only group and a ddH2O group served as controls). The combination was centrifuged, and the supernatant was transferred to a 96-well plate. The absorbance at 540 nm was measured to evaluate the extent of haemolysis.
Characterization of PMVs@PLGA complexes in vivo
The metabolism of PMVs@PLGA complexes was examined utilizing wholesome SD rats to analyse the final sample of their metabolism. For biotoxicity evaluation, venous blood was collected from wholesome SD rats inside 1–10 days after injection of PMVs@PLGA complexes and analysed for neutrophil share utilizing a completely automated haematology analyser (BC-5000 Vet, Mindray, China), liver and kidney perform markers utilizing a completely automated biochemical analyser (Vetube 30, Mindray, China), myoglobin (MYO) focus utilizing a Rat MYO ELISA Package (E-EL-R0053c, Elabscience, China), a Rat Troponin I Kind 3 (cTnI) ELISA Package (E-EL-R1253c, Elabscience, China) for the cTnI focus, and a Rat CKMB (Creatine Kinase MB Isoenzyme) ELISA Package (E-EL-R1327c, Elabscience, China) for the CK-MB focus. For focusing on validation, rats have been used to assemble a MIRI mannequin after which injected with DiR-labelled PMVs, PLGA and PMVs@PLGA complexes analysis by reside imaging. Moreover, coronary heart tissue was collected from the rats to make frozen sections, and the toxicity of the PMVs@PLGA complexes was once more verified utilizing TUNEL apoptosis evaluation and immunofluorescence (IF) evaluation of BAX and BCL-2 expression.
In vivo imaging
For the metabolic profile assay, PLGA was labelled with DiR and assembled into PMVs@PLGA complexes, suspended in PBS and injected into wholesome SD rats through the tail vein. The guts, liver, spleen, lungs and kidneys have been collected at 7 time factors, 0, 2, 4, 6, 12, 24 and 48 h postinjection, respectively, after which rinsed appropriately with PBS, and pictures have been taken utilizing an animal reside imaging system (ABL X5, Tanon, China) and analysed for common photons per pixel per ms (common PPP (per ms)).
For the focusing on profile assay, PMVs, PLGA and PMVs@PLGA complexes have been labelled with DiR and injected into SD rats through the tail vein 10 min after MIRI mannequin development. Twenty-four hours later, the guts, liver, spleen, lungs and kidneys of the rats have been collected and washed appropriately with PBS, and pictures have been taken utilizing an animal reside imaging system (ABL X5, Tanon, China) and analysed for common photons per pixel per ms (common PPP (per ms)).
Stream cytometry assay
For the cell particle uptake effectivity assay, PMVs have been labelled with Dil, and PLGA was labelled with FITC and assembled into PMVs@PLGA complexes. A complete of 20 μg of PMVs, 20 μg of PLGA, or 20 μg of PMVs assembled with 20 μg of PLGA was added to at least one effectively of a 6-well plate in 2 mL of medium and incubated with H9C2 cells for twenty-four h. The H9C2 cells have been collected and washed 3 occasions with precooled PBS, the cell focus was adjusted to 1 × 106 cells/mL, and the particle uptake effectivity was measured utilizing a move cytometer (FACSCalibur, BD, USA).
For the apoptosis assay, handled H9C2 cells have been collected, washed 3 times with precooled PBS and stained with an Annexin V-Alexa Fluor 647/PI Apoptosis Assay Package (FMSAV647, Fcmacs, China) in line with the producer’s directions, adopted by move cytometry (FACSCalibur, BD, USA) to analyse the cells for apoptosis.
TUNEL apoptosis assay
The TUNEL apoptosis assay was carried out utilizing a One-step TUNEL In Situ Apoptosis Package (E-CK-A322, Elabscience, China). For cell samples, after 4% paraformaldehyde fixation and 0.3% Triton-X100 permeabilization, the TUNEL staining working resolution was configured for staining in line with the producer’s directions, and the nuclei have been stained utilizing DAPI staining resolution, washed appropriately with PBS and noticed with a fluorescence microscope. For frozen sections of tissue, after 4% paraformaldehyde fixation and proteinase-Okay permeabilization, the TUNEL staining working resolution was configured for staining in line with the producer’s directions, and the nuclei have been stained utilizing DAPI staining resolution, washed appropriately with PBS and noticed with a fluorescence microscope.
Caspase-3 exercise assay
The caspase-3 exercise assay was carried out utilizing a GreenNuc Caspase-3 Assay Package for Dwell Cells (C1168 M, Beyotime, China). After staining handled reside cells with GreenNuc staining resolution configured in line with the producer’s directions, they have been fastened utilizing 4% paraformaldehyde and permeabilized with 0.3% Triton X-100. The nuclei have been subsequently stained utilizing Hoechst staining resolution and noticed utilizing fluorescence microscopy.
qPCR
Complete RNA was remoted from cells utilizing Invitrogen TRIzol reagent (15596026, Thermo Fisher Scientific, USA). For mRNA evaluation, a reverse transcription response was carried out through the RevertAid First Strand cDNA Synthesis Package (K1622, Thermo Fisher Scientific) in line with the producer’s directions. qPCR was carried out utilizing PowerUp SYBR Inexperienced Grasp Combine (A25742, Thermo Fisher Scientific, USA) on a QuantStudio 5 Actual-Time PCR System (A28569, Thermo Fisher Scientific, USA). All goal genes have been normalized to GAPDH. The primer sequences used are listed as follows:
rno-Nrf2 ahead, 5′-TCCAAGTCCAGAAGCCAAACTGAC-3′;
rno-Nrf2 reverse, 5′-GGAGAGGATGCTGAAGGAATC-3′;
rno-GAPDH ahead, 5′-GACATGCCGCCTGGAGAAAC-3′;
rno-GAPDH reverse, 5′-AGCCCAGGATGCCCTTTAGT-3′.
For miRNA evaluation, a reverse transcription response was carried out through an EZ-press microRNA Reverse Transcription Package (miRT2-L, EZBioscience, USA) in line with the producer’s directions. qPCR was carried out utilizing an EZ-press microRNA qPCR Package (ROX2 plus) (miQP2, EZBioscience, USA) on a QuantStudio 5 Actual-Time PCR System. All goal genes have been standardized to U6. The primer sequences used are listed as follows:
rno-miR-144-3p ahead, 5′-GCGCGCGTACAGTATAGATGA-3′;
rno-miR-155-5p ahead, 5′-GCGCGTTAATGCTAATTGTGAT-3′;
rno-miR-140-5p ahead, 5′-CGCGCAGTGGTTTTACCCTA-3′;
rno-miR-153-3p ahead, 5′-CGCGTTGCATAGTCACAAAA-3′;
rno-miR-410-3p ahead, 5′-CGCGGCAATTTAGTGTGTGT-3′;
rno-miR-27a-3p ahead, 5′-TTCACAGTGGCTAAGTTCCGC-3′;
rno-miR-27b-3p ahead, 5′-TTCACAGTGGCTAAGTTCTGC-3′;
rno-miR-340-5p ahead, 5′-GCGCGTTATAAAGCAATGAGA-3′;
rno-miR-106b-5p ahead, 5′-TAAAGTGCTGACAGTGCAGAT-3′;
rno-miR-495 ahead, 5′-AAACAAACATGGTGCACTTCTT-3′;
rno-miR-142-5p ahead, 5′-GCGCGCATAAAGTAGAAAGC-3′;
rno-miR-17-5p ahead, 5′-CAAAGTGCTTACAGTGCAGGTAG-3′;
rno-miR-93-5p ahead, 5′- CAAAGTGCTGTTCGTGCAGGTAG-3′;
rno-miR-128-3p ahead, 5′-CGCGTCACAGTGAACCGGT-3′;
U6 ahead, 5’-CCTGCTTCGGCAGCACA-3’.
The reverse primers for the miRNAs and U6 have been offered within the EZ-press microRNA qPCR Package (ROX2 plus). Quantification of qPCR outcomes was carried out by the two−ΔΔCt methodology.
Transfection of miRNA mimic, inhibitor and antagomir
For in vitro transfection, one effectively of a 6-well plate was used. After the cells grew to 60–80% confluence, 20 μg of PMVs, 20 μg of PLGA and 200 pmol of miRNA in 125 μL of basal medium have been assembled into PMVs@PLGA-miRNA complexes and blended totally and gently. The cells have been allowed to face for 15 min after which added to the whole medium within the 6-well plate to finish the transfection.
For in vivo transfection, every rat was ready with antagomir at 10 mg/kg for transfection. The corresponding weight of antagomir was made into PMVs@PLGA-miRNA antagomir in vitro. Roughly 10 min after the completion of the MIRI mannequin, it was injected into the rat through the tail vein to finish the transfection.
Twin-luciferase reporter gene assays
HEK-293T cells have been seeded right into a 24-well plate, incubated in a single day, after which transfected with the twin luciferase plasmid vectors and miR-140-5p, miR-144-3p, miR-155-5p, and miR-340-5p mimics. In response to the producer’s protocol, the luciferase exercise was checked by a FLUOROSKAN FL SYSTEM (5200220, Thermo Fisher Scientific, USA) utilizing the Twin-Luciferase Reporter Assay System (E1910, Promega, USA). The firefly luciferase exercise was normalized to the Renilla luciferase exercise in every effectively.
Western blot (WB) evaluation and immunofluorescence (IF) evaluation
Cell pellets or tissue homogenates have been lysed with RIPA lysis buffer (WB3100, NCM Biotech, China) and PMSF (P0100, Solarbio, China) for 1 h; after centrifugation at 12,000 rpm for 15 min, the supernatant was aspirated, blended with loading buffer, and incubated at 100 °C for 10 min to acquire protein samples. Western blot (WB) experiments have been carried out utilizing SDS‒PAGE. The first antibodies used have been anti-Nrf2 (1:1000) (T55136, Abmart, China), anti-β-actin (1:10,000) (AC026, ABclonal, China), CD31 (1:1000) (11,265–1-AP, Proteintech, China), anti-CD41 (1:1000) (18,308–1-AP, Proteintech, China), anti-CD42b (1:1000) (12,860–1-AP, Proteintech, China), and anti-CD61 (1:1000) (18,309–1-AP, Proteintech, China). The secondary antibody was HRP-conjugated goat anti-rabbit IgG (1:10,000) (RS0002, Immunoway, USA). Proteins have been detected by a chemiluminescent imaging system (ChemDoc MP, BIO-RAD, USA), and greyscale evaluation was carried out utilizing ImageJ software program.
Frozen sections have been used for immunofluorescence (IF). After 0.3% Triton X-100 permeabilization and 5% BSA blocking, the slides have been incubated in a single day with anti-Nrf2 (1:100) (T55136, Abmart, China), anti-BAX (1:100) (50,599-2-Ig, Proteintech, China), and anti-BCL-2 (1:100) (26,593-1-AP, Proteintech, China) antibodies at 4 °C. The sections have been incubated with the secondary antibody CoraLite594-conjugated goat anti-rabbit IgG (1:500) (SA00013-4, Proteintech, China) at RT for 1 h, and the nuclei have been stained with DAPI (C1002, Beyotime, China).
Triphenyl tetrazolium chloride (TTC) staining
After mannequin preparation was accomplished or therapy was accomplished, the guts was eliminated, rinsed in saline, sliced into 2 mm sections and positioned in 2% TTC resolution (G3005, Solarbio, China) and incubated at 37 °C at midnight for 30 min. The guts sections have been then fastened in 4% paraformaldehyde for six h, photographed after which analysed for the dimensions of the white areas.
Statistical evaluation
The info have been evaluated by GraphPad Prism software program and expressed because the imply ± SD. Variations between teams have been assessed by one-way evaluation of variance and subsequent Tukey posttests. P < 0.05 signifies significance.