Dec 13, 2023
(Nanowerk Information) The commentary of biomolecular buildings utilizing atomic power microscopy (AFM) and the direct visualization of purposeful conformational dynamics in high-speed AFM (HS-AFM) experiments have considerably superior the understanding of organic processes on the nanoscale. In experiments, a organic pattern is deposited on a supporting floor (AFM substrate) and is scanned by a probing tip to detect the molecular form and its dynamical adjustments.
The commentary of protein dynamics underneath HS-AFM is a fragile stability between immobilizing the construction on the supporting floor whereas on the identical time stopping too robust perturbations by immobilization.
The method of inserting a biomolecular pattern on the supporting floor and controlling its correct attachment is a problem on the very begin of each AFM commentary. By the chemical composition of the buffer, interactions between the pattern and substrate may be modified. Such floor modifications are sometimes essential for profitable AFM observations of protein buildings and their purposeful motions. Nevertheless, the molecular orientation of the pattern is a priori unknown, and attributable to limitations within the spatial decision of photos, troublesome to deduce from a posteriori evaluation.
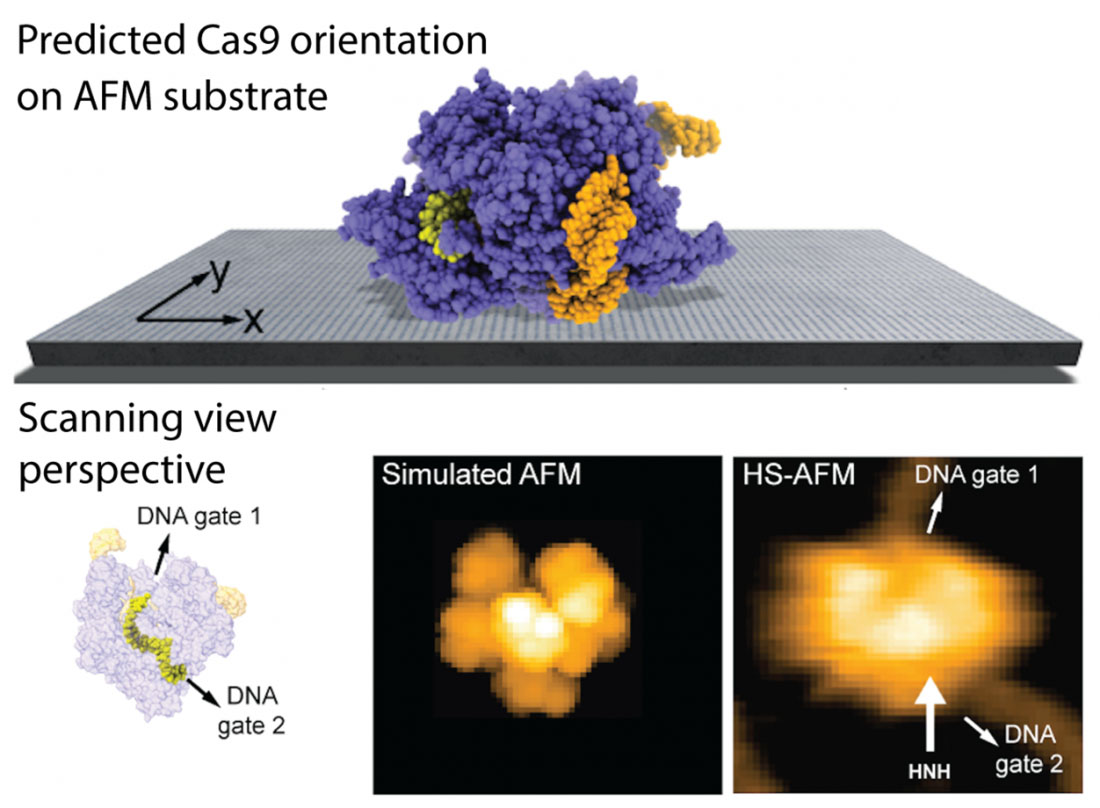 Fig. 1: Instance utility of the developed methodology. The anticipated orientation of the Cas9 protein on the AFM substrate is proven, and a simulated AFM picture calculated within the scanning view perspective exhibits exceptional settlement with earlier HS-AFM imaging. HS-AFM can reliably visualize purposeful relative motions of goal DNA and Cas9 as a result of the entry and exit gates of the DNA strand, which is situated in a tunnel inside the Cas9 construction, aren’t blocked by contacts with the APTES-mica substrate. (Picture: Amyot, Nakamoto, Kodera and Flechsig)
Romain Amyot, Noriyuki Kodera, and Holger Flechsig from Kanazawa College have now developed a bodily mannequin to foretell the location of biomolecular buildings on AFM substrates based mostly on electrostatic interactions. The tactic considers the substrates generally utilized in AFM experiments (mica, APTES-mica, lipid bilayers) and takes into consideration buffer situations. In pc simulations, a lot of attainable molecular preparations on the AFM substrate are sampled, and from evaluating the corresponding interplay energies, probably the most favorable placement is set. Moreover, the evaluation permits predictions of the imaging stability underneath tip scanning.
The group printed their findings in Frontiers in Molecular Biosciences (“Predicting the location of biomolecular buildings on AFM substrates based mostly on electrostatic interactions”).
The authors present a number of purposes of the brand new methodology and procure exceptional settlement of mannequin predictions with earlier experimental HS-AFM imaging of proteins. The findings can clarify, for instance, why HS-AFM observations of the Cas9 endonuclease, a protein taking part in a key position in genetic engineering purposes, can reliably visualize purposeful relative motions of goal DNA and Cas9 and seize DNA cleavage occasions on the single molecule stage (see Fig. 1). Moreover, as demonstrated for the ATP-powered chaperone machine ClpB, the mannequin can clarify how buffer situations have an effect on the steadiness of the sample-substrate advanced and validate observations of earlier HS-AFM experiments.
In abstract, the brand new methodology permits to make use of the large quantity of obtainable structural knowledge for biomolecules to make predictions of the pattern placement on AFM substrates even previous to an precise experiment, and it will also be utilized for post-experimental evaluation of AFM imaging knowledge. The developed methodology is carried out inside the freely accessible BioAFMviewer software program bundle, offering a handy platform for purposes by the broad BioAFM neighborhood.
Fig. 1: Instance utility of the developed methodology. The anticipated orientation of the Cas9 protein on the AFM substrate is proven, and a simulated AFM picture calculated within the scanning view perspective exhibits exceptional settlement with earlier HS-AFM imaging. HS-AFM can reliably visualize purposeful relative motions of goal DNA and Cas9 as a result of the entry and exit gates of the DNA strand, which is situated in a tunnel inside the Cas9 construction, aren’t blocked by contacts with the APTES-mica substrate. (Picture: Amyot, Nakamoto, Kodera and Flechsig)
Romain Amyot, Noriyuki Kodera, and Holger Flechsig from Kanazawa College have now developed a bodily mannequin to foretell the location of biomolecular buildings on AFM substrates based mostly on electrostatic interactions. The tactic considers the substrates generally utilized in AFM experiments (mica, APTES-mica, lipid bilayers) and takes into consideration buffer situations. In pc simulations, a lot of attainable molecular preparations on the AFM substrate are sampled, and from evaluating the corresponding interplay energies, probably the most favorable placement is set. Moreover, the evaluation permits predictions of the imaging stability underneath tip scanning.
The group printed their findings in Frontiers in Molecular Biosciences (“Predicting the location of biomolecular buildings on AFM substrates based mostly on electrostatic interactions”).
The authors present a number of purposes of the brand new methodology and procure exceptional settlement of mannequin predictions with earlier experimental HS-AFM imaging of proteins. The findings can clarify, for instance, why HS-AFM observations of the Cas9 endonuclease, a protein taking part in a key position in genetic engineering purposes, can reliably visualize purposeful relative motions of goal DNA and Cas9 and seize DNA cleavage occasions on the single molecule stage (see Fig. 1). Moreover, as demonstrated for the ATP-powered chaperone machine ClpB, the mannequin can clarify how buffer situations have an effect on the steadiness of the sample-substrate advanced and validate observations of earlier HS-AFM experiments.
In abstract, the brand new methodology permits to make use of the large quantity of obtainable structural knowledge for biomolecules to make predictions of the pattern placement on AFM substrates even previous to an precise experiment, and it will also be utilized for post-experimental evaluation of AFM imaging knowledge. The developed methodology is carried out inside the freely accessible BioAFMviewer software program bundle, offering a handy platform for purposes by the broad BioAFM neighborhood.
 Fig. 1: Instance utility of the developed methodology. The anticipated orientation of the Cas9 protein on the AFM substrate is proven, and a simulated AFM picture calculated within the scanning view perspective exhibits exceptional settlement with earlier HS-AFM imaging. HS-AFM can reliably visualize purposeful relative motions of goal DNA and Cas9 as a result of the entry and exit gates of the DNA strand, which is situated in a tunnel inside the Cas9 construction, aren’t blocked by contacts with the APTES-mica substrate. (Picture: Amyot, Nakamoto, Kodera and Flechsig)
Romain Amyot, Noriyuki Kodera, and Holger Flechsig from Kanazawa College have now developed a bodily mannequin to foretell the location of biomolecular buildings on AFM substrates based mostly on electrostatic interactions. The tactic considers the substrates generally utilized in AFM experiments (mica, APTES-mica, lipid bilayers) and takes into consideration buffer situations. In pc simulations, a lot of attainable molecular preparations on the AFM substrate are sampled, and from evaluating the corresponding interplay energies, probably the most favorable placement is set. Moreover, the evaluation permits predictions of the imaging stability underneath tip scanning.
The group printed their findings in Frontiers in Molecular Biosciences (“Predicting the location of biomolecular buildings on AFM substrates based mostly on electrostatic interactions”).
The authors present a number of purposes of the brand new methodology and procure exceptional settlement of mannequin predictions with earlier experimental HS-AFM imaging of proteins. The findings can clarify, for instance, why HS-AFM observations of the Cas9 endonuclease, a protein taking part in a key position in genetic engineering purposes, can reliably visualize purposeful relative motions of goal DNA and Cas9 and seize DNA cleavage occasions on the single molecule stage (see Fig. 1). Moreover, as demonstrated for the ATP-powered chaperone machine ClpB, the mannequin can clarify how buffer situations have an effect on the steadiness of the sample-substrate advanced and validate observations of earlier HS-AFM experiments.
In abstract, the brand new methodology permits to make use of the large quantity of obtainable structural knowledge for biomolecules to make predictions of the pattern placement on AFM substrates even previous to an precise experiment, and it will also be utilized for post-experimental evaluation of AFM imaging knowledge. The developed methodology is carried out inside the freely accessible BioAFMviewer software program bundle, offering a handy platform for purposes by the broad BioAFM neighborhood.
Fig. 1: Instance utility of the developed methodology. The anticipated orientation of the Cas9 protein on the AFM substrate is proven, and a simulated AFM picture calculated within the scanning view perspective exhibits exceptional settlement with earlier HS-AFM imaging. HS-AFM can reliably visualize purposeful relative motions of goal DNA and Cas9 as a result of the entry and exit gates of the DNA strand, which is situated in a tunnel inside the Cas9 construction, aren’t blocked by contacts with the APTES-mica substrate. (Picture: Amyot, Nakamoto, Kodera and Flechsig)
Romain Amyot, Noriyuki Kodera, and Holger Flechsig from Kanazawa College have now developed a bodily mannequin to foretell the location of biomolecular buildings on AFM substrates based mostly on electrostatic interactions. The tactic considers the substrates generally utilized in AFM experiments (mica, APTES-mica, lipid bilayers) and takes into consideration buffer situations. In pc simulations, a lot of attainable molecular preparations on the AFM substrate are sampled, and from evaluating the corresponding interplay energies, probably the most favorable placement is set. Moreover, the evaluation permits predictions of the imaging stability underneath tip scanning.
The group printed their findings in Frontiers in Molecular Biosciences (“Predicting the location of biomolecular buildings on AFM substrates based mostly on electrostatic interactions”).
The authors present a number of purposes of the brand new methodology and procure exceptional settlement of mannequin predictions with earlier experimental HS-AFM imaging of proteins. The findings can clarify, for instance, why HS-AFM observations of the Cas9 endonuclease, a protein taking part in a key position in genetic engineering purposes, can reliably visualize purposeful relative motions of goal DNA and Cas9 and seize DNA cleavage occasions on the single molecule stage (see Fig. 1). Moreover, as demonstrated for the ATP-powered chaperone machine ClpB, the mannequin can clarify how buffer situations have an effect on the steadiness of the sample-substrate advanced and validate observations of earlier HS-AFM experiments.
In abstract, the brand new methodology permits to make use of the large quantity of obtainable structural knowledge for biomolecules to make predictions of the pattern placement on AFM substrates even previous to an precise experiment, and it will also be utilized for post-experimental evaluation of AFM imaging knowledge. The developed methodology is carried out inside the freely accessible BioAFMviewer software program bundle, offering a handy platform for purposes by the broad BioAFM neighborhood.


