On this article, we are going to learn to construct a classifier utilizing a easy Convolution Neural Community which might classify the pictures of affected person’s xray to detect wheather the affected person is Regular or affected by Pneumonia.
To get extra understanding, observe the steps accordingly.
Importing Libraries
The libraries we are going to utilizing are :
- Pandas- The pandas library is a well-liked open-source knowledge manipulation and evaluation software in Python. It supplies an information construction referred to as a DataFrame, which is analogous to a spreadsheet or a SQL desk, and permits for straightforward manipulation and evaluation of knowledge.
- Numpy- NumPy is a well-liked open-source library in Python for scientific computing, particularly for working with numerical knowledge. It supplies instruments for working with massive, multi-dimensional arrays and matrices, and provides a variety of mathematical capabilities for performing operations on these arrays.
- Matplotlib- It’s a well-liked open-source knowledge visualization library in Python. It supplies a spread of instruments for creating high-quality visualizations of knowledge, together with line plots, scatter plots, bar plots, histograms, and extra.
- TensorFlow- TensorFlow is a well-liked open-source library in Python for constructing and coaching machine studying fashions. It was developed by Google and is broadly utilized in each academia and trade for quite a lot of functions, together with picture and speech recognition, pure language processing, and suggestion programs.
Python3
|
|
Importing Dataset
To run the pocket book within the native system. The dataset will be downloaded from [ https://www.kaggle.com/datasets/paultimothymooney/chest-xray-pneumonia ]. The dataset is within the format of a zipper file. So to import after which unzip it, by operating the beneath code.
Python3
|
|
Learn the picture dataset
On this part, we are going to attempt to perceive visualize some pictures which have been supplied to us to construct the classifier for every class.
Let’s load the coaching picture
Python3
|
|
Output:
['PNEUMONIA', 'NORMAL']
This reveals that, there are two courses that we’ve right here i.e. Regular and Pneumonia.
Python3
|
|
Output:
There are 3875 pictures of pneumonia imfected in coaching dataset There are 1341 regular pictures in coaching dataset
Plot the Pneumonia contaminated Chest X-ray pictures
Python3
|
|
Output:
.png)
Pneumonia contaminated Chest X-ray pictures
Plot the Regular Chest X-ray pictures
Python3
|
|
Output:
.png)
Regular Chest X-ray pictures
Information Preparation for Coaching
On this part, we are going to classify the dataset into practice,check and validation format.
Python3
|
|
Output:
Discovered 5216 information belonging to 2 courses. Discovered 624 information belonging to 2 courses. Discovered 16 information belonging to 2 courses.
Mannequin Structure
The mannequin structure will be described as follows:
- Enter layer: Conv2D layer with 32 filters, 3×3 kernel measurement, ‘relu’ activation perform, and enter form of (256, 256, 3)
- MaxPooling2D layer with 2×2 pool measurement
- Conv2D layer with 64 filters, 3×3 kernel measurement, ‘relu’ activation perform
- MaxPooling2D layer with 2×2 pool measurement
- Conv2D layer with 64 filters, 3×3 kernel measurement, ‘relu’ activation perform
- MaxPooling2D layer with 2×2 pool measurement
- Conv2D layer with 64 filters, 3×3 kernel measurement, ‘relu’ activation perform
- MaxPooling2D layer with 2×2 pool measurement
- Flatten layer
- Dense layer with 512 neurons, ‘relu’ activation perform
- BatchNormalization layer
- Dense layer with 512 neurons, ‘relu’ activation perform
- Dropout layer with a charge of 0.1
- BatchNormalization layer
- Dense layer with 512 neurons, ‘relu’ activation perform
- Dropout layer with a charge of 0.2
- BatchNormalization layer
- Dense layer with 512 neurons, ‘relu’ activation perform
- Dropout layer with a charge of 0.2
- BatchNormalization layer
- Output layer: Dense layer with 2 neurons and ‘sigmoid’ activation perform, representing the possibilities of the 2 courses (pneumonia or regular)
In abstract our mannequin has :
- 4 Convolutional Layers adopted by MaxPooling Layers.
- Then One Flatten layer to obtain and flatten the output of the convolutional layer.
- Then we may have three absolutely linked layers adopted by the output of the flattened layer.
- We’ve got included some BatchNormalization layers to allow secure and quick coaching and a Dropout layer earlier than the ultimate layer to keep away from any chance of overfitting.
- The ultimate layer is the output layer which has the activation perform sigmoid to categorise the outcomes into two courses(i,e Regular or Pneumonia).
Python3
|
|
Print the abstract of the mannequin structure:
Output:
Mannequin: "sequential"
_________________________________________________________________
Layer (sort) Output Form Param #
=================================================================
conv2d (Conv2D) (None, 254, 254, 32) 896
max_pooling2d (MaxPooling2D (None, 127, 127, 32) 0
)
conv2d_1 (Conv2D) (None, 125, 125, 64) 18496
max_pooling2d_1 (MaxPooling (None, 62, 62, 64) 0
2D)
conv2d_2 (Conv2D) (None, 60, 60, 64) 36928
max_pooling2d_2 (MaxPooling (None, 30, 30, 64) 0
2D)
conv2d_3 (Conv2D) (None, 28, 28, 64) 36928
max_pooling2d_3 (MaxPooling (None, 14, 14, 64) 0
2D)
flatten (Flatten) (None, 12544) 0
dense (Dense) (None, 512) 6423040
batch_normalization (BatchN (None, 512) 2048
ormalization)
dense_1 (Dense) (None, 512) 262656
dropout (Dropout) (None, 512) 0
batch_normalization_1 (Batc (None, 512) 2048
hNormalization)
dense_2 (Dense) (None, 512) 262656
dropout_1 (Dropout) (None, 512) 0
batch_normalization_2 (Batc (None, 512) 2048
hNormalization)
dense_3 (Dense) (None, 512) 262656
dropout_2 (Dropout) (None, 512) 0
batch_normalization_3 (Batc (None, 512) 2048
hNormalization)
dense_4 (Dense) (None, 2) 1026
=================================================================
Complete params: 7,313,474
Trainable params: 7,309,378
Non-trainable params: 4,096
_________________________________________________________________
The enter picture we’ve taken initially resized into 256 X 256. And later it remodeled into the binary classification worth.
Plot the mannequin structure:
Python3
|
|
Output:
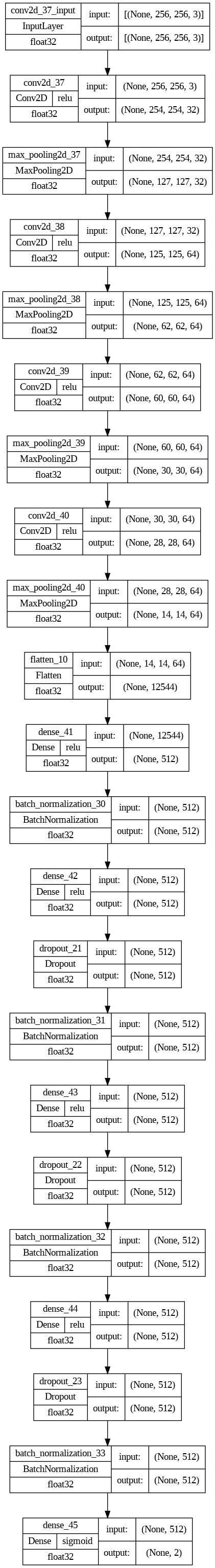
Mannequin
Compile the Mannequin:
Python3
|
|
Prepare the mannequin
Now we are able to practice our mannequin, right here we outline epochs = 10, however you possibly can carry out hyperparameter tuning for higher outcomes.
Python3
|
|
Output:
Epoch 1/10 163/163 [==============================] - 59s 259ms/step - loss: 0.2657 - accuracy: 0.9128 - val_loss: 2.1434 - val_accuracy: 0.5625 Epoch 2/10 163/163 [==============================] - 34s 201ms/step - loss: 0.1493 - accuracy: 0.9505 - val_loss: 3.0297 - val_accuracy: 0.6250 Epoch 3/10 163/163 [==============================] - 34s 198ms/step - loss: 0.1107 - accuracy: 0.9626 - val_loss: 0.5933 - val_accuracy: 0.7500 Epoch 4/10 163/163 [==============================] - 33s 197ms/step - loss: 0.0992 - accuracy: 0.9640 - val_loss: 0.3691 - val_accuracy: 0.8125 Epoch 5/10 163/163 [==============================] - 34s 202ms/step - loss: 0.0968 - accuracy: 0.9651 - val_loss: 3.5919 - val_accuracy: 0.5000 Epoch 6/10 163/163 [==============================] - 34s 199ms/step - loss: 0.1012 - accuracy: 0.9653 - val_loss: 3.8678 - val_accuracy: 0.5000 Epoch 7/10 163/163 [==============================] - 34s 198ms/step - loss: 0.1026 - accuracy: 0.9613 - val_loss: 3.2006 - val_accuracy: 0.5625 Epoch 8/10 163/163 [==============================] - 35s 204ms/step - loss: 0.0785 - accuracy: 0.9701 - val_loss: 1.7824 - val_accuracy: 0.5000 Epoch 9/10 163/163 [==============================] - 34s 198ms/step - loss: 0.0717 - accuracy: 0.9745 - val_loss: 3.3485 - val_accuracy: 0.5625 Epoch 10/10 163/163 [==============================] - 35s 200ms/step - loss: 0.0699 - accuracy: 0.9770 - val_loss: 0.5788 - val_accuracy: 0.6250
Mannequin Analysis
Let’s visualize the coaching and validation accuracy with every epoch.
Python3
|
|
Output:
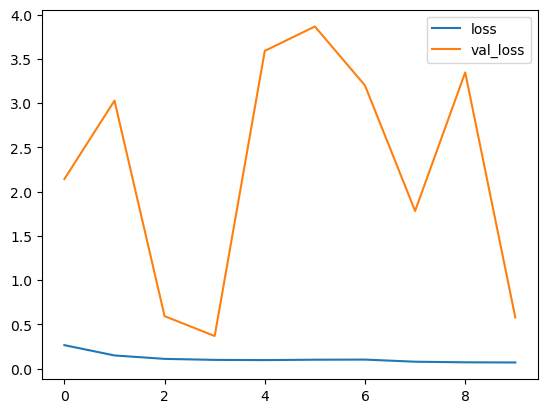
Losses per iterations
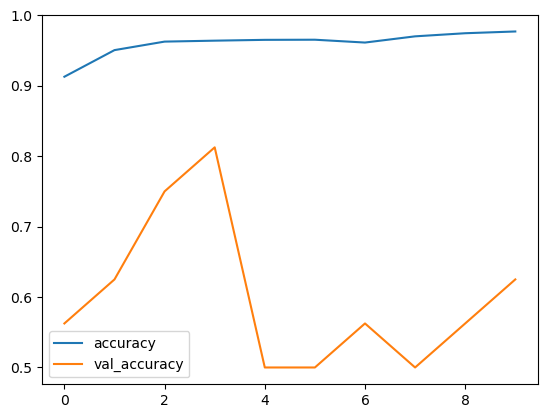
Accuracy per iterations
Our mannequin is performing good on coaching dataset, however not on check dataset. So, that is the case of overfitting. This can be as a result of imbalanced dataset.
Discover the accuracy on Take a look at Datasets
Python3
|
|
Output:
20/20 [==============================] - 4s 130ms/step - loss: 0.4542 - accuracy: 0.8237 The accuracy of the mannequin on check dataset is 82.0
Prediction
Let’s verify the mannequin for random pictures.
Python3
|
|
Output:
1/1 [==============================] - 0s 328ms/step Pneumonia
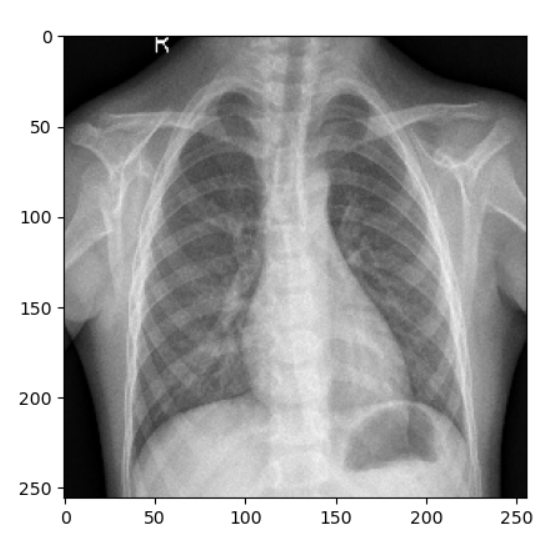
Regular Chest X-ray
Python3
|
|
Output:
1/1 [==============================] - 0s 328ms/step Pneumonia
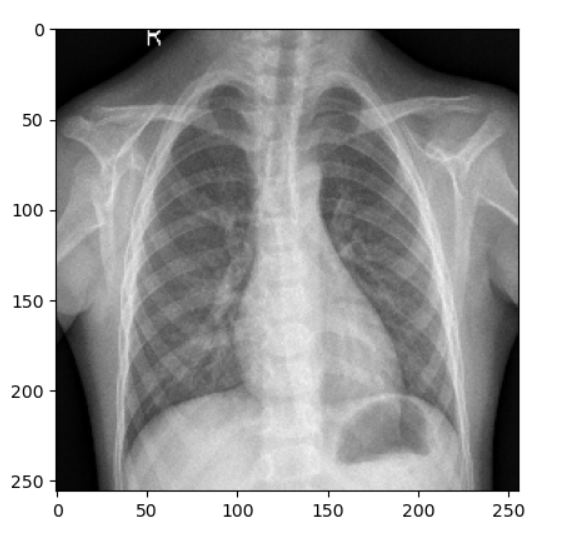
Pneumonia Contaminated Chest
Conclusions:
Our mannequin is performing effectively however as per losses and accuracy curve per iterations. It’s overfitting. This can be as a result of unbalanced dataset. By balancing the dataset with an equal variety of regular and pneumonia pictures. We will get a greater consequence.


