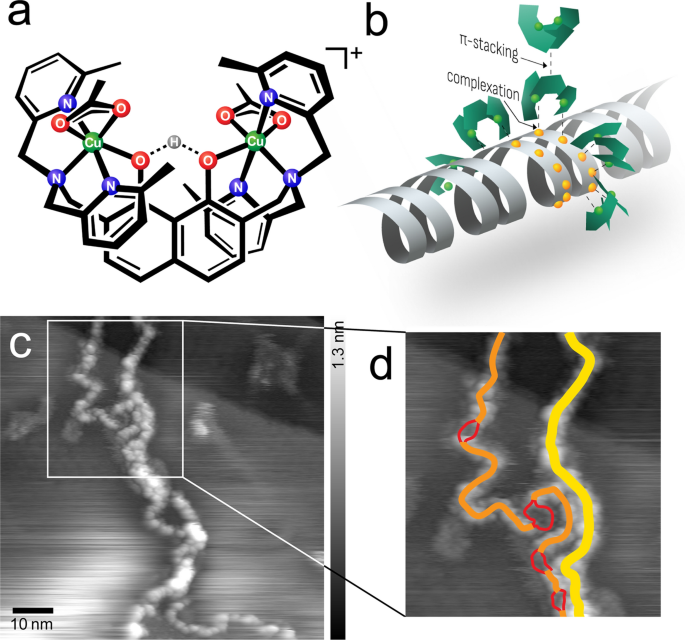
Determine 1a, b illustrate the design of the [(HtomMe){Cu(OAc)}2]+ complexes, with two steel cores which can be positioned by the naphthalene spine at a distance of 0.6–0.7 nm, offering the exact molecular recognition potential of two neighboring phosphate moieties of the DNA spine. Every of the 2 pyridyl teams coordinate across the steel atoms in such a manner that they sterically restrict the hydrolytic cleavage response and hamper the nicking of the strand [24].
a Chemical construction of the [(HtomMe){Cu(OAc)}2]+ complexes. The descriptor for the complexes modifications based on the chemical environment, for clarification see Further file 1: Determine S1. b Mannequin of binding modes of the advanced molecules to adsorb to DNA and to one another. c UHV-AFM topographic picture of the agglomeration of a number of strands (frequency shift setpoint df =− 2.5 Hz, oscillation amplitude A = 14.1 nm, resonance frequency f0 = 259.0 Hz) and the devoted interpretation d: two double strands (yellow, top 0.6 nm), one singular ds (orange, top 0.4 nm), melting bubble dissociation in two single strands (crimson, top 0.1 nm)
An answer of λ-phage DNA answer (Lambda DNA Hind III Digest, Sigma-Aldrich, 12.5 ng/µl) (λ-DNA) was handled with a 0.4 nM answer of the dinuclear copper complexes [(HtomMe){Cu(OH2)}2]3+, with an incubation time of 45 h, as this was decided to show an indicated onset of anticipated results to the DNA. Concerning the focus dependent affect of the complexes to DNA, a dilution sequence will be discovered within the Further File 1: (Determine S2).
Bottom bonding
A outcome that was already anticipated from investigations below ambient circumstances was the interstrand agglomeration [4]. The high-resolution photographs taken below UHV circumstances in Fig. 1c, d present how a number of strands align in an effort to agglomerate, which will be geometrically traced to particular person strands.
As Jany et al. already speculated, this impact outcomes from π-stacking results of the outwards-oriented naphthalene und pyridyl residue moieties of [(HtomMe){Cu(OAc)}2]+. X-ray diffraction confirmed that the freely rotatable pyridyl teams orient in such a manner that, along with the middle naphthalene, they kind three parallel ridges, which might permit two molecules to connect by coercing a number of π-stacking interactions.
Melting bubbles
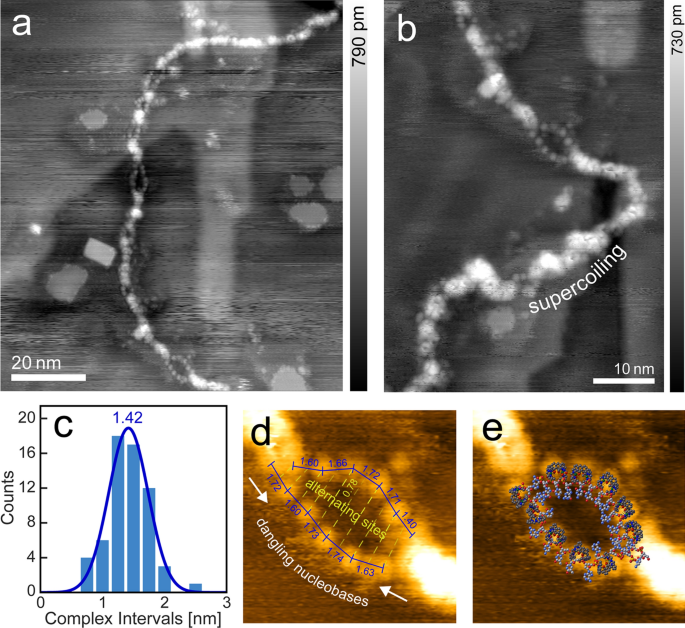
One other exceptional function is launched in Fig. 2. Alongside the strand seem a number of partial fissions of varied size into thinner strands, with these sectors ranging in size from 1 to eight nm. They don’t seem to be solely characterised by a decreased top (0.1–0.2 nm in distinction to 0.4 nm of the principle strand), but in addition by a string of bead-like look with equidistant protrusions alongside a strand.
a Dissociated melting bubbles alongside a double strand of λ-DNA (taken below UHV, df =− 2.0 Hz, A = 14.1 nm, f0 = 259.0 Hz). b Nearer have a look at one of many bigger bubbles, together with an space of supercoiled buildup. c Distribution of interval distances between the protrusion options alongside the bubbles, with a imply of 1.42 nm. d Distances and antiphasic patterning of options. The bigger than common distance of 0.78 nm will be attributed to observations the place advanced coated ssDNA areas seem elongated [25]. e Overlay of a doable molecular mannequin to match the noticed topography [DNA model from [26] (1BNA)]
The phenomenon of sectoral dehybridization of DNA double strands, referred to as melting-, denaturation- or respiration bubbles, is well-known in DNA science [27]. It’s essential to DNA replication, by introducing insertion level for single strand interacting polymerase enzymes [28, 29] and is usually related to AT-rich (adenine, thymine) DNA areas, that are, as a result of two as an alternative of three hydrogen bonds, barely much less strongly certain than their GC-counterparts [30] (guanine, cytosine).
They had been noticed not directly (by fluorescence quenching [31, 32]) and instantly (TEM [25] and AFM [33]), primarily in round DNA and plasmids, the place the openings are induced by twist stress. They seem additionally in linear DNA [34], precipitated statistically by molecular movement [27, 35,36,37].
The peak of the measured protrusions lies with 0.2 nm nicely beneath the one measured for a double strand in air (1 nm) and vacuum (0.4 nm). As well as, the typical interval of the protrusions is with 1.42 nm too giant for a single nucleobase size and too low for a double helix pitch, however properly matches the twofold distance between two neighboring phosphate teams. Subsequently, we conclude that the “melted bubble” sections certainly encompass dehybridized double DNA strands into two single strands, the place the beads representing the coordinatively bonded [(HtomMe)Cu2]3+ molecules. It shall even be famous that upon nearer inspection, a zigzag sort association of antiphasic binding websites alongside the only strands can readily be discerned (see Fig. 2d).
Reasoning of the melting bubbles
With the dissociated sections making up roughly 12% of the contour size of a strand, which is even above the same old prevalence charge of single strand areas in linear DNA of 4% in supercoiled kind [25], the remark of the formation right here doesn’t look like a purely statistical course of.
Melting bubbles don’t seem steadily below ambient circumstances and are clearly not attributable to the UHV circumstances, tough dealing with or freezing as extra measurements have proven. To the very best of our data, it has not been proven in literature the place DNA adsorbed to low adhesion substrates was subjected to hoover [38, 39].
Since they don’t structurally change over time, it can be excluded that they’re the results of tip-induced injury by the scanning movement. Subsequently, a correlation to the presence of the metal-coordinated spine binding advanced molecules is understandable.
DNA habits in vacuum
To appropriately assess these findings, it’s important to have in mind the habits of dsDNA in vacuum surroundings, in addition to the affect of the substrate.
It’s broadly recognized that dsDNA in environments of low humidity transitions right into a extra densely packed formation, the so-called A-form. That is attributable to the decrease availability of stabilizing water molecules and ions surrounding the negatively charged spine. A decrease helical pitch distance and a extra pronounced main groove are attribute for the A-form [40, 41].
Since it’s the commonest selection of substrate for investigations below ambient circumstances, most AFM-works of DNA in vacuum circumstances function the DNA adsorbed onto functionalized mica. This closely influences the structural properties of the adsorbed strands, whether or not it conserves the physiological B-form [15] or modifications the elastical properties derived from persistence lengths and equilibrated geometries [42]. The reason being that in an effort to connect DNA to mica, it’s crucial to make use of a functionalization, to keep away from the repelling destructive DNA fees of the phosphate spine in addition to of the mica (({textual content{SiO}}_{x}^{-})). Subsequently, totally different strategies are employed, that provide totally different affinities to the DNA. This ranges from much less invasive approaches by utilizing divalent cations [12] like Mg2+ or Ni2+, to the usage of sturdy bonding floor modifications like APTES [42], ODA [34], PLL [43] or PLO [44]. The power of the bonding influences the noticed geometry, for the reason that stronger the bonding, the much less dynamics is allowed after adsorption. This results in 2D equilibrated strands on ionic functionalization and 3D projected strands on covalent bonding surfaces. Electrically impartial substrates like Au or HOPG provide little to no attraction to DNA, disregarding mirror-dipoles and related van-der-Waals forces, thus, making them largely unsuitable to be used below ambient and liquid circumstances. In UHV then again, gold is of nice benefit, since it may be ultra-cleanly ready and atomically managed in-situ, whereas mica introduces a definite roughness that’s extremely disadvantageous to high-resolution measurements.
Since literature is sparse, relating to the interplay of DNA with gold, its habits throughout electrospray ionization and even its habits in vacuum circumstances, an applicable sequence of checks of deposition on totally different substrates has been performed.
Gold
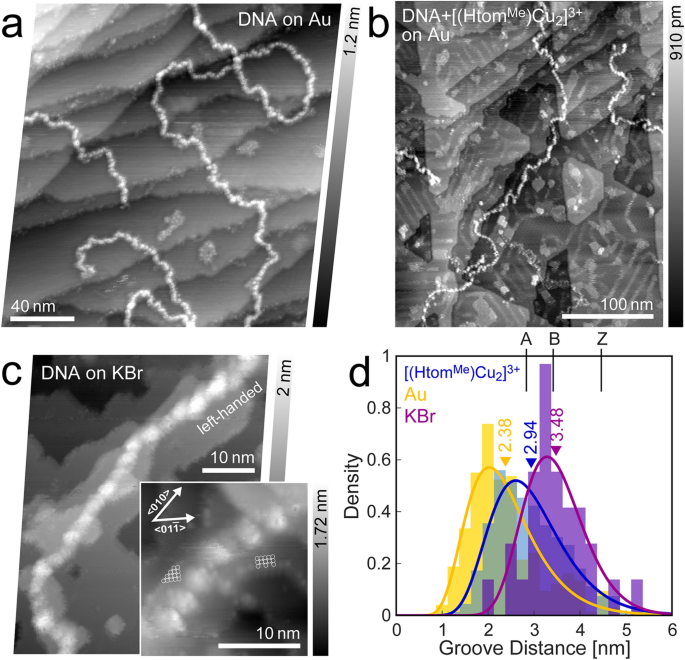
A string of bead-like look characterizes the pristine DNA-strands on gold (Fig. 3a). The grooves between the options correlate to the foremost groove [38] whereas smaller options couldn’t be resolved, as a result of instability of the pattern. One other clarification options for the beads to be correlated to electrostatic forces from residual ions, that are recognized to reside primarily within the grooves [45, 46]. Attributable to the low interplay between strand and substrate, the strands are simply moved throughout scanning, which prevents larger decision. Determine 3d reveals a distribution of measured groove distances. The imply worth of two.38 nm is in settlement to the anticipated pitch worth vary for A-form reworked DNA (2.82 nm [47]). The related strand top measures to about 0.5 nm.
λ-DNA subjected to UHV circumstances on totally different substrates. a Untreated DNA on Au(111) (df =− 6.2 Hz, A = 12.6 nm, f0 = 286.3 Hz). b [(HtomMe)Cu2]3+-treated DNA on Au(111) (df =− 2.0 Hz, A = 14.1 nm, f0 = 259.0 Hz). The residues on the in any other case flat atomic planes are probably as a result of residual advanced molecules. c DNA on potassium bromide (df =− 8.6 Hz, A = 4.7 nm, f0 = 302.0 Hz, inset: df =− 20.2 Hz, A = 1.9 nm, f0 = 302.0 Hz). d Distribution of groove distances, together with corresponding imply values for various substrates (Au, KBr)
Potassium bromide
As a reference, pristine λ-DNA strands had been moreover deposited on an in-situ cleaved floor of KBr. Strands can clearly be recognized, with options that resemble the foremost groove, even regardless of indicators of minor rearrangement of ion plateaus, probably attributable to residual solvent hitting the floor (Fig. 3c).
A statistical evaluation of groove distances reveals excessive settlement to the anticipated full flip distance of three.4 nm of the B-form DNA-configuration.
For many elements, the right-handedness will be readily noticed. Nevertheless, upon nearer look, segments of differing groove distance and even left-handedness, resembling A- and even the Z-form of dsDNA [41], will be moreover discerned. That is recognized to happen below excessive salt concentrations at GC-rich sections [48] and even on Ni-functionalized mica [49].
The anticipated vacuum-induced transition to the A-form is outwardly hampered for essentially the most half, as a result of an abundance of ions that act stabilizing to the physiological B-form and having the ability to retain a commensurate variety of water molecules from evaporating [45].
The measured dsDNA top is about 0.6 nm, as a result of flattening from adhesion forces and embedding into residual solvent.
[(Htom
Me
)Cu
2
]
3+
-bound DNA
Other than the already mentioned strand aggregations and bubble formations, the DNA bonded with [(HtomMe)Cu2]3+ reveals an identical string of bead-like look, away from the bigger aggregations (Fig. 3b). A statistical evaluation of their groove distances results in a imply worth of two.94 nm. Whereas the pitch can barely be influenced by the bottom sequence [48], that is far in between the untreated DNA on gold that has taken on the A-form and the presumably conserved B-form on KBr.
There have been no variations in persistence lengths noticeable for every of the three instances, residing every time round 10 nm.