Respiratory tract infections have been acknowledged to be one of many key causes of demise worldwide. A number of viral infections manifest related signs, and clinically differentiating between them is difficult. Nonetheless, correct prognosis of those infections is extraordinarily essential to supply the perfect therapy to the sufferers.
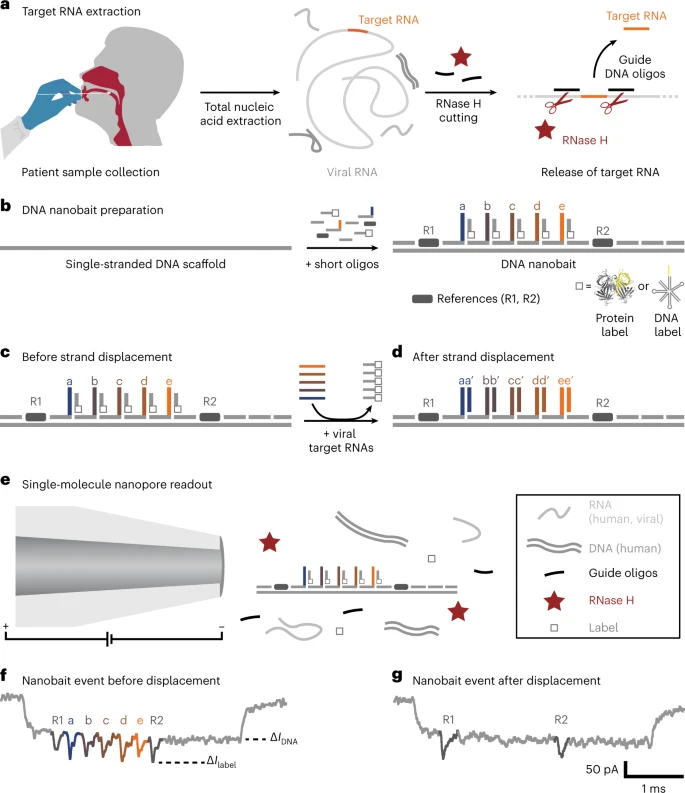
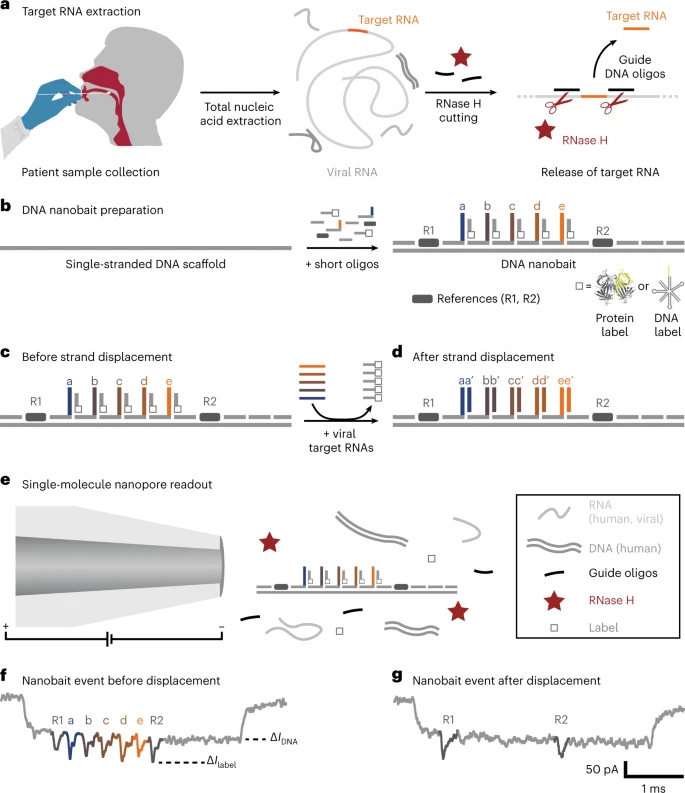
Self-assembled DNA nanobait technique for multiplexed viral diagnostics. a, Oropharyngeal swab pattern is collected from sufferers suspected to have COVID-19. The full nucleic acids had been extracted (human and viral RNAs are proven in mild gray; DNA is proven in darkish gray), and the goal RNA was cleaved out utilizing programmable RNase H chopping. Such a handled pattern was additional examined for viral presence. b, Nanobait is made by mixing a single-stranded DNA scaffold (M13mp18 DNA) with quick oligonucleotides the place a few of them carry complementary seize strands (a–e) for particular targets (a’–e’) in a goal viral RNA. As well as, {a partially} complementary oligo with a structural label (protein or DNA primarily based; white sq.) is added to every website to amplify the sign in nanopore recordings. We marked a sensing area with two references (darkish gray). c,d, Nanobait earlier than (c) and after (d) strand displacement reactions of 5 targets (colored strands). If the targets are current, the 5 gray strands with labels are displaced. The 2 outer alerts originate from reference constructions that point out the sensing area, and the 5 binding websites between the references are particular for the 5 completely different targets. e, Every nanobait is voltage pushed by the nanopore and detected in a mix of molecules. f, Typical nanopore present signature as a perform of time for a nanobait as designed with 5 labels current. The primary present drop corresponds to DNA (ΔIDNA) and the second to labels (ΔIlabel). g, Typical nanobait occasion after strand displacement of all of the 5 targets. The presence of targets is detected by the lacking downward alerts particular to every goal. Picture Credit score: Bošković, F. et al. (2023), Nature Nanotechnology
Not too long ago, researchers at The College of Cambridge have focussed on creating an progressive methodology primarily based on DNA nanobait, which might concurrently detect a number of respiratory viruses and their variants in lower than an hour. This research is obtainable in Nature Nanotechnology.
Challenges in Respiratory An infection Analysis
Round 65% of viral infections are incorrectly identified as pneumonia, and roughly 95% of misdiagnosed sufferers obtain pointless antimicrobial therapies. The present coronavirus illness 2019 (COVID-19) pandemic, brought on by the fast outbreak of extreme acute respiratory syndrome coronavirus-2 (SAR-CoV-2) virus, additional underscores the diagnostic limitations of quickly figuring out the virus variants.
As a consequence of genomic mutations, the continuous emergence of SARS-CoV-2 variants has extended the pandemic regardless of the supply of COVID-19 vaccines. This virus has claimed thousands and thousands of lives because of the lack of fast diagnostic measures. In response to Professor Stephen Baker from the Cambridge Institute of Therapeutic Immunology and Infectious Illness, who co-authored the current research, “For sufferers, we all know that fast prognosis improves their final result, so with the ability to detect the infectious agent shortly might save their life,”.
A few of the strategies used to determine viral variants are quantitative reverse transcription–polymerase chain response (qRT-PCR), adopted by molecular sequencing. Though polymerase chain response (PCR)-based diagnostic strategies are generally used to determine nucleic acids from advanced organic samples, they’ve restricted multiplexing capabilities. Subsequently, there may be an pressing want for a novel diagnostic methodology to detect viruses in addition to their variants in diminished pattern quantity.
Not too long ago, nanotechnology strategies, reminiscent of nanopore sensing, have been utilized for virus detection. This method can differentiate between a number of nucleic acid species primarily based on the uniquely engineered DNA nanostructure. These DNA nanostructures have the potential for multiplexed biosensing.
Improvement and Working Precept of DNA Nanobait
Scientists aimed to beat the aforementioned diagnostic limitations utilizing a nanotechnology-based method. This methodology relies on a DNA nanobait, which might concurrently detect a number of respiratory viruses and their variants. The detection methodology relies on acquiring swab samples of sufferers, adopted by nucleic acid extraction and programmable RNase H chopping of viral RNA.
On this approach, the RNA targets are predominantly chosen through oligonucleotides, a single-stranded DNA, which might bind at a particular area (each upstream and downstream) of the viral genome. Subsequently, RNase H was employed to digest the RNA sequence in RNA: DNA hybrids, and launch the center goal RNA, which was recognized utilizing sequence-specific binding to the nanobait.
The genetically engineered DNA nanobait contained 5 binding websites, which might bind to as many RNA targets. The nanobait was assembled by combing a single-stranded DNA scaffold with a number of quick complementary oligonucleotides. One finish of the nanobait contained a sensing area, a DNA overhang complementary to the goal sequence. A correct nanobait meeting was confirmed by electrophoretic mobility shift assay and atomic power microscopy (AFM) imaging instruments.
A goal sequence binds to certainly one of six unpaired bases by changing the oligonucleotide with the label at its complementary overhang. This methodology is named toehold-mediated strand displacement. Subsequently, the presence of the goal nucleotide was detected by the absence of a label on the respective website.
A nanopore DNA sensor detects the nanobait primarily based on its voltage-driven translocation. The negatively charged nanobait travels, by a slender opening, in direction of a positively charged electrode in an electrolyte resolution, eliciting a definite present blockage signature. Every nanobait–nanopore occasion reveals the presence of RNA targets. The distinctive nanobait designs enabled the identification of targets originating from numerous elements of the identical virus or quite a few viral genomes.
Simultaneous Identification of A number of Viral Variants
The newly developed nanobait will help determine a number of respiratory viruses, reminiscent of a respiratory syncytial virus (RSV), SARS-CoV-2, parainfluenza, and influenza. The authors demonstrated that the newly designed DNA nanobait might detect RSV primarily based on site-specific displacement.
The induction of spikes after nanopore occasions indicated the absence of targets, which confirmed the presence of particular targets for RSV, SARS-CoV-2, rhinovirus, parainfluenza, and influenza. Importantly, the 5 websites of nanobait enabled simultaneous identification of wild-type SARS-CoV-2 and its 4 variants, particularly, B.1, B.1.1.7 (Alpha), B.1.351 (Beta), and B.1.617 (Delta).
The DNA nanobait was programmed utilizing a single-nucleotide mismatch, which enabled the identification of the SARS-CoV-2 unique pressure and newly emerged variants. Notably, in a nanopore sensor, dark-colored bars had been noticed for the unique SARS-CoV-2 pressure and light-colored bars for all 4 variants.
The developmental process of the DNA nanobaits is extraordinarily advantageous as a result of it skips preamplification and purification steps, that are doubtlessly time-intensive and costly steps.
Whereas discussing the longer term software of the nanobait, Professor Ulrich Keyser of Cavendish Laboratory, who additionally co-authored the research, stated, “Nanobait relies on DNA nanotechnology and can enable for a lot of extra thrilling purposes sooner or later,” He additional added, “For business purposes and roll-out to the general public we must convert our nanopore platform right into a hand-held gadget.”
Reference
Bošković, F. et al. (2023) Simultaneous identification of viruses and viral variants with programmable DNA nanobait. Nature Nanotechnology. https://doi.org/10.1038/s41565-022-01287-x.
Supply: College of Cambridge